Open Access Article
Open Access ArticleStrategies to overcome antibiotic resistance: silver nanoparticles and vancomycin in pathogen eradication
Sakshi V.
Khairnar
 a,
Ashish
Das
a,
Ashish
Das
 b,
David
Oupický
a,
Marat
Sadykov
b and
Svetlana
Romanova
b,
David
Oupický
a,
Marat
Sadykov
b and
Svetlana
Romanova
 *a
*a
aCenter for Drug Delivery and Nanomedicine, Department of Pharmaceutical Sciences, University of Nebraska Medical Center, Omaha, NE 68198, USA. E-mail: sromanova@unomaha.edu
bDepartment of Pathology, Microbiology and Immunology, University of Nebraska Medical Center, Omaha, NE 68198, USA
First published on 20th February 2025
Abstract
The increasing prevalence of antibiotic resistance presents a significant challenge to public health, undermining the efficacy of conventional antibiotic treatments. Given the scarcity of new antibiotics and efficient preventive strategies, the exploration of alternative treatments has become imperative. For many years, vancomycin, a glycopeptide antibiotic, has been considered a last resort for treating severe Gram-positive bacterial infections. However, the emergence of vancomycin-resistant bacteria has raised significant concerns. The expanding use of nanomaterials in healthcare settings has shifted the spotlight towards innovative antibacterial nanomaterials, potentially offering solutions to the resistance crisis. One of the promising approaches to combat resistance involves employing metal nanoparticles to enhance antibiotic efficacy. Silver nanoparticles (AgNPs) have garnered particular interest due to their extensively documented broad-spectrum and robust antimicrobial properties, especially against bacterial biofilms, making them useful against multidrug-resistant pathogens. Recent evidence suggests synergistic antibacterial activity when AgNPs are combined with vancomycin. This innovative approach offers the potential to mitigate associated side effects and improve susceptibility to resistant strains. Consequently, the combination of vancomycin and AgNPs presents a compelling strategy for addressing bacterial infections. This review delves into the interactions between AgNPs and vancomycin, providing valuable insights into combating antibiotic resistance. Current research efforts continue to investigate and underscore the advancement of formulation strategies and their performance evaluation in a wide array of infection paradigms. This continuing work aims to enhance our understanding of drug delivery systems and their therapeutic potential across various infectious diseases.
1. Introduction
Antimicrobial resistance refers to the ability of microorganisms to resist or endure the effects of antimicrobial drugs, including commonly used antibiotics. Bacteria can develop drug resistance through intrinsic traits or acquired mechanisms, rendering antibiotics less effective.1,2 In the United States, it is estimated that annually, more than 2.8 million individuals acquire antimicrobial-resistant infections, leading to over 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities.3 Future projections suggest that deaths due to antimicrobial resistance will increase over the next few decades, with a nearly 70% rise by 2050 compared to 2022, predominantly affecting the aging population.4 The prevalence of multidrug-resistant (MDR) bacteria, which exhibit resistance to at least one agent in three or more antimicrobial classes, has become increasingly common, particularly in healthcare settings.5 In July 2024, the Centers for Disease Control and Prevention (CDC) reported a 20% increase in six bacterial antimicrobial-resistant hospital-onset infections during the COVID-19 pandemic. These infections peaked in 2021 and remained above pre-pandemic levels in 2022.6 Concerns have been raised about the ‘post-antibiotic era’, where common infections may become life-threatening due to ineffective treatments. Many experts, including those at the CDC, believe we are already experiencing this era.3 In 2017, the World Health Organization (WHO) released its first-ever list of “priority pathogens” in light of increasing antibiotic resistance. It identified 12 families of bacteria associated with increased disease burden and treatment failures.7,8 Bacteria can develop resistance through several mechanisms, including the destruction or modification of antibiotics, prevention of access to the target, mutations in target genes or acquisition of genes encoding drug resistance, alteration of the antibiotic target site, and resistance due to global cellular adaptive processes, rendering antibiotics ineffective (Fig. 1).2
000 fatalities.3 Future projections suggest that deaths due to antimicrobial resistance will increase over the next few decades, with a nearly 70% rise by 2050 compared to 2022, predominantly affecting the aging population.4 The prevalence of multidrug-resistant (MDR) bacteria, which exhibit resistance to at least one agent in three or more antimicrobial classes, has become increasingly common, particularly in healthcare settings.5 In July 2024, the Centers for Disease Control and Prevention (CDC) reported a 20% increase in six bacterial antimicrobial-resistant hospital-onset infections during the COVID-19 pandemic. These infections peaked in 2021 and remained above pre-pandemic levels in 2022.6 Concerns have been raised about the ‘post-antibiotic era’, where common infections may become life-threatening due to ineffective treatments. Many experts, including those at the CDC, believe we are already experiencing this era.3 In 2017, the World Health Organization (WHO) released its first-ever list of “priority pathogens” in light of increasing antibiotic resistance. It identified 12 families of bacteria associated with increased disease burden and treatment failures.7,8 Bacteria can develop resistance through several mechanisms, including the destruction or modification of antibiotics, prevention of access to the target, mutations in target genes or acquisition of genes encoding drug resistance, alteration of the antibiotic target site, and resistance due to global cellular adaptive processes, rendering antibiotics ineffective (Fig. 1).2
Many strategies have been devised to combat drug-resistant bacteria, including bacteriophage therapy, antimicrobial peptides, phytochemicals, metallo-antibiotics, and combination therapies.9 Metal nanoparticles have emerged as a promising avenue for research, demonstrating significant antibacterial activity against resistant strains.10 Among them, silver nanoparticles (AgNPs) offer the potential to address these challenges due to their broad-spectrum and robust antimicrobial properties, especially against bacterial biofilms.11,12 While planktonic bacteria are susceptible, biofilms, which are far more resistant to antibiotics, account for nearly 80% of all microbial infections in the body.13 Some reports suggest biofilm formation can prevent the penetration of drugs, increasing the effective doses up to 1000-fold compared to planktonic bacteria.14 Therefore, combining conventional antibiotics with AgNPs offers an effective strategy for enhancing antibacterial efficacy.15 AgNPs influence, induce, and modulate different inflammatory, metabolic, biochemical, and cellular processes, contributing to their multifaceted antimicrobial action in addressing multidrug resistance in bacteria. Their direct antimicrobial activity is linked to adhesion to cell surfaces, penetration, intracellular damage, induction of oxidative stress by reactive oxygen species (ROS) generation, and modulation of signal transduction pathways.16
This review explores the potential of AgNPs in combination with vancomycin, the first discovered glycopeptide antibiotic. Despite its effectiveness against a wide range of Gram-positive bacteria, vancomycin has notable limitations. These include its poor stability, short half-life in vivo, substantial first-pass effect, slow bactericidal activity, poor tissue penetration (especially in the lungs), and the risk of severe side effects like nephrotoxicity and ototoxicity.17,18 Its high molecular weight and hydrophilic nature result in poor gastrointestinal absorption, limiting its administration to intravenous routes, which are associated with patient compliance issues and systemic toxicity.19 While nanocarrier systems, such as polymeric micelles and surface-modified liposomes, improve drug delivery, they face challenges in stability during storage, scalable production, and regulatory concerns over long-term safety and immunogenicity.20 Furthermore, variability in patient responses, driven by immune status and comorbidities, further complicates treatment outcomes, highlighting the urgent need for novel and effective therapeutic strategies.
Investigating the combination of AgNPs and vancomycin presents a promising approach to tackle the growing challenge of vancomycin-resistant Gram-positive bacteria. The conjugation of vancomycin with AgNPs has demonstrated enhanced antibacterial activity against Gram-positive bacteria, including Vancomycin-resistant Enterococci (VRE) and methicillin-resistant Staphylococcus epidermidis (MRSE).21 This synergistic effect not only expands the range of treatable infections but also improves vancomycin's ability to penetrate biofilms, thereby enhancing its bactericidal action. Such combination therapy may offer a more effective solution for managing infections that are increasingly difficult to treat with traditional antibiotics alone.
2. Silver nanoparticles
The bactericidal activity of silver has been documented since ancient times. It is effective against over 650 microorganisms and provides a means to control vector-borne infections.22,23 In the early 17th century, Angelus Sola utilized systemic silver nitrate to treat epilepsy and cholera. Subsequently, during the 19th century, German obstetrician Carl Crede and Austrian surgeon Johann Nepomuk Rust used a diluted solution of silver nitrate to prevent microbial infections.24 Given the increasing decline in antibiotic effectiveness, AgNPs have regained prominence, with the Food and Drug Administration (FDA) approving numerous silver-infused wound dressings and topical agents for marketing.25,26 In wound management, for example, a clinical trial by Pathi et al. demonstrated that Kadermin, a silver nanoparticle-based cream, outperformed Mupirocin in promoting faster wound healing and bacterial clearance. This highlights the potential for AgNPs to serve as antibiotic-sparing agents in treating infections.27 While AgNPs are capable of killing both Gram-positive and Gram-negative bacteria, they demonstrate higher efficacy against Gram-negative bacteria,16 as shown in Table 1.| Bacteria | Size (nm) | MIC (μg mL−1) | Type of strain | Ref. |
|---|---|---|---|---|
| Abbreviation: minimum inhibitory concentration (MIC). | ||||
| Acinetobacter baumannii | 10–50 | 4–25 | Gram-negative | 28 |
| Burkholderia pseudomallei | 10–20 | 32–48 | Gram-negative | 29 |
| Campylobacter jejuni | 10–50 | 4.92–39.4 | Gram-negative | 30 |
| Escherichia coli | 5 | 10 | Gram-negative | 31 |
| Enterobacter hormaechei | 5–15 | 4 | Gram-negative | 32 |
| Helicobacter pylori | 10–40 | 5 | Gram-negative | 33 |
| Klebsiella pneumonia | 50–65 | 62.5–125 | Gram-negative | 34 |
| Neisseria gonorrhoeae | 120 | 12.5 | Gram-negative | 35 |
| Pseudomonas aeruginosa | 45–53 | 2 | Gram-negative | 36 |
| Proteus mirabilis | 54 | 10 | Gram-negative | 37 |
| Salmonella typhus | 5–45 | 16 | Gram-negative | 38 |
| Shigella flexneri | 40–170 | 20 | Gram-negative | 39 |
| Vibrio cholera | 20–30 | 10 | Gram-negative | 40 |
| Yersinia enterocolitica | 16.56 | 50 | Gram-negative | 41 |
| Bacillus subtilis | 5–55 | 0.8–6 | Gram-positive | 42 |
| Clostridium difficile | 25–35 | — | Gram-positive | 43 |
| Enterococcus faecalis | — | 5 | Gram-positive | 44 |
| Listeria monocytogenes | 2–35 | 8 | Gram-positive | 45 |
| Micrococcus luteus | 62.09 | 60 | Gram-positive | 46 |
| Mycobacterium tuberculosis | 3–20 | 12.5 | Gram-positive | 47 |
| Staphylococcus epidermidis | 10–100 | 1–6 | Gram-positive | 48 |
| Staphylococcus aureus | 11.02–17.92 | 4–64 | Gram-positive | 49 |
| Streptococcus mutans | 10–78 | 4–16 | Gram-positive | 50 |
Recent studies have explored the application of AgNPs in dental materials such as adhesives, root canal fillers, implant surfaces, and denture resins due to their ability to inhibit bacterial growth and biofilm formation. Ongoing research is investigating their potential as nano-vectors for gene transfer and drug delivery, highlighting the expanding role of AgNPs in advancing oral health treatments.51 Moreover, AgNPs are being investigated for therapeutic applications beyond antimicrobial uses. Rehman et al. demonstrated that AgNPs synthesized from plant extracts exhibit notable antidiabetic properties.52 In another study, Khajeh et al. showed that AgNPs derived from the extracts of Moringa oleifera could upregulate the MLH1 (Mult homolog1) gene, which plays a crucial role in promoting DNA repair and apoptosis in colorectal cancer cells.53 AgNPs are also being explored for their antiviral activity against a wide range of viruses, including hepatitis B virus, human immunodeficiency virus, herpes simplex virus, and monkeypox virus, indicating their ability to inhibit viral attachment and entry into host cells.54 This multifunctionality paves the way for AgNP applications beyond healthcare, making it a highly sought-after material across multiple industries. The antibacterial potential of AgNPs has led to their incorporation into a variety of consumer and medical products, including textiles,55 agriculture,56 packaging,57 optoelectronics,58 and environmental remediation.59
Despite significant progress, the widespread clinical and industrial adoption of AgNPs remains restricted by challenges such as cytotoxicity, environmental concerns, scalability, manufacturing uniformity, and regulatory constraints.60,61 Additionally, achieving precise control over nanoparticle size, morphology, and dispersion is a persistent hurdle.62 Addressing these factors will be crucial in ensuring the safe and effective utilization of AgNPs in biomedical applications and expanding their commercial viability.
2.1. Synthesis of AgNPs
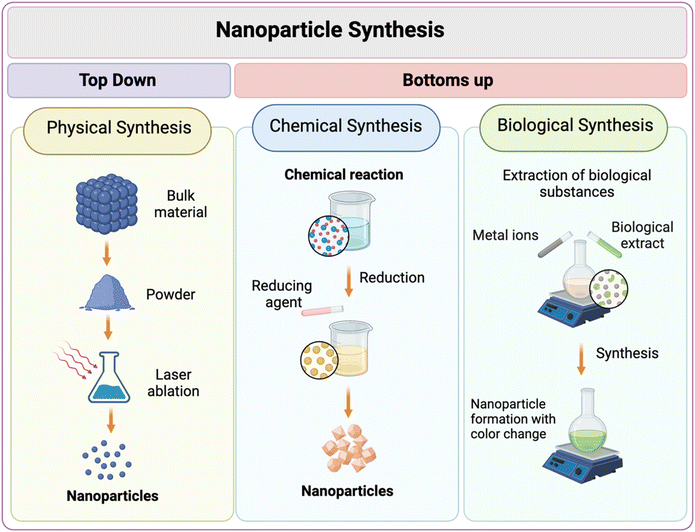
AgNPs can be synthesized through a variety of methods, which include both modern biochemical techniques and traditional approaches. These methods provide the flexibility to control the size and structural properties of the nanoparticles. Researchers employ diverse techniques, broadly categorized into top-down and bottom-up approaches, as illustrated in Fig. 2, to produce AgNPs that meet specific requirements.63 The top-down approach involves breaking down bulk material into nanoparticles, utilizing techniques such as laser ablation, evaporation–condensation, and mechanical milling. This approach offers the advantage of producing pure and uncontaminated AgNPs without relying on hazardous chemicals, ensuring the production of high-quality nanoparticles. However, it is energy-intensive and requires advanced machinery, significant space, and long processing times, often resulting in relatively low yields. Conversely, bottom-up approaches are cost-effective, simple, and produce high yields with controllable particle sizes. However, they often involve the use of toxic reducing agents and stabilizers, leading to harmful waste, residual contaminants, and potential aggregation of nanoparticles.64,65 This approach involves building nanoparticles from individual atoms or molecules in solution, encompassing methods like sol–gel processes and chemical reduction. The latter typically requires a silver salt precursor, reducing agents, and stabilizing chemicals.66–68 Notably, the use of stabilizing or capping agents is crucial in this process. These agents are often employed to enhance stability and prevent coagulation.69 This is particularly important because nanoparticles, due to their significantly reduced size compared to bulk materials, possess higher surface energy, which can lead to instability.70 These agents work through various mechanisms, commonly categorized into five types: steric stabilization, electrostatic stabilization, depletion stabilization, stabilization by hydration forces, and stabilization by Van der Waals forces.54Biological synthesis methods, a subset of bottom-up approaches, present a more eco-friendly and cost-effective alternative, utilizing plant extracts or microorganisms as reducing agents. This method leverages the reducing and stabilizing power of natural compounds, including flavonoids, alkaloids, and enzymes from plants or microorganisms, to control nanoparticle formation and enhance stability without introducing harsh chemicals.71 Recent studies have demonstrated their effectiveness against various pathogens, including Candida albicans, E. coli, and S. aureus, often surpassing traditional antibiotics in efficacy. For instance, AgNPs synthesized using Bacillus methylotrophicus exhibited superior inhibition of microbial growth compared to conventional antibiotics.72 Another study utilized Aspergillus sydowii to produce AgNPs with a spherical shape and sizes varying between 1 to 24 nm. These nanoparticles demonstrated significant antiproliferative and antifungal activities against clinical fungal pathogens, suggesting their potential use in biomedical applications.73 Additionally, plant-based synthesis methods, such as those using Prunus yedoensis, have shown promise in producing AgNPs with enhanced effectiveness against skin bacteria compared to commercial alternatives.74 Palanisamy et al. reviewed marine biomolecule-assisted synthesis using compounds such as polysaccharides, enzymes, vitamins, and proteins derived from a wide range of marine organisms, including but not limited to seaweed, seagrass, and microalgae.75 This growing interest in biological synthesis is driven not only by its reduced environmental impact but also by its cost-effectiveness and reliability, positioning it as an attractive method for producing AgNPs to combat antibiotic resistance.66 However, challenges remain, such as ensuring reproducibility, optimizing large-scale production, and understanding the long-term effects of AgNPs in vivo.
2.2. Physicochemical properties of AgNPs
The physicochemical properties of AgNPs, such as size, shape, surface charge, and surface functionalization, play a critical role in their antimicrobial efficacy and interactions with both bacteria and host tissues. Zaman et al. highlighted the crucial influence of AgNP size on photocatalytic and antibacterial applications. Smaller nanoparticles exhibit enhanced efficacy against bacterial strains due to their higher surface-area-to-volume ratio and ability to penetrate bacterial cell barriers. Additionally, they demonstrated greater activity against Gram-negative bacterial strains.76 Previous research has shown challenges in synthesizing highly monodisperse and stable AgNPs below 10 nm or above 50 nm using single reducing agents. Strong reducers like sodium borohydride (NaBH4) produce small, uniform particles but struggle with larger sizes, while weaker reducers like trisodium citrate (TSC) yield larger particles with wider size distributions and varied shapes. A co-reduction method using both NaBH4 and TSC may offer better control over nucleation and growth, potentially addressing these limitations.77In addition to size, the shape of AgNPs significantly influences their antibacterial effectiveness. AgNPs can be synthesized in various shapes, including spherical, triangular, cubic, rod-shaped, platelet, hexagonal, pyramidal, flower-shaped, octahedral, tetrahedral, cylindrical, and irregular configurations, each exhibiting unique physicochemical properties.78,79 In a study, truncated-triangular nanoparticles appeared to be more effective for microbial killing. However, spherical nanoparticles are still considered to be the best-suited for practical applications in either colloidal form or immobilized state.77,80 However, it's important to note that other factors also play crucial roles in determining antibacterial efficacy. Pokhrel et al. investigated the antibacterial efficacy of 5 nm positively charged amine (NH2)-functionalized AgNPs and 45 nm negatively charged citrate–AgNPs against E. coli. Positively charged AgNPs demonstrate greater antibacterial activity due to electrostatic attraction with negatively charged bacterial cell membranes.81 Functionalization can prevent AgNP aggregation, enhancing their stability in various solvents. For example, poly(pentafluorostyrene) (PPFS)-modified AgNPs showed improved dispersion in organic solvents, while glycosylated PPFS-modified AgNPs were more easily dispersed in aqueous medium.82 Surface modifications enable the targeted delivery of AgNPs to specific tissues or organs while potentially reducing toxicity concerns. Functionalizing AgNPs with biomolecules such as antibiotics, polymers, glycolipids, amino acids, and peptides enhances their stability, biocompatibility, and functionality for diverse applications.83
Depending on the desired local or systemic effect, AgNPs can be administered through topical applications or via intravenous injection.84,85 He et al. developed an Ag-functionalized chitosan hydrogel dressing with sustained silver release and enhanced antibacterial capabilities. The hydrogel demonstrated significant wound healing acceleration, achieving a 99% healing rate in a mouse model, with improved re-epithelialization and reduced inflammation.86 While effective against various conditions, AgNP cytotoxicity remains a concern for widespread use in medical applications. Studies are focusing on addressing safety concerns, optimizing efficacy, and developing novel applications to harness the full potential of AgNPs in biomedicine while ensuring their safe use.
2.3. Mechanism of antibacterial activity of AgNPs
While the broad-spectrum antibacterial properties of AgNPs are well-known, the precise mechanisms behind their antimicrobial effects remain unclear. The antibacterial effectiveness of AgNPs mainly arises from the release of Ag+ ions. These positively charged ions engage in electrostatic interactions with the negatively charged functional groups containing phosphorous and sulfur, such as phosphates (–PO43−) or sulfates (–SO42−) in cellular membranes, proteins, and DNA bases.87 The presence of Ag+ ions can enhance the permeability of the cytoplasmic membrane, potentially causing the bacterial envelope to rupture. Once the free Ag+ ions are internalized, they can inhibit respiratory enzymes within the electron transport chain (ETC) and leading to the generation of ROS.88 This oxidative stress can damage DNA and interrupt the metabolic activities of enzymes containing iron–sulfur clusters, such as aconitase in the tricarboxylic acid cycle (TCA), through Fenton chemistry.89,90 Furthermore, Ag+ ions can compromise the DNA's ability to replicate and disrupt protein synthesis by denaturing ribosomes in the cytoplasm.91 It has also been noticed that AgNPs have antibacterial properties without the release of ions. AgNPs tend to accumulate in the pits that form on the cell wall. Subsequently, they attach to the cell membrane surface, altering its permeability by changing the cell potential and hindering cellular respiration.92 The ability of AgNPs to interfere with bacterial signal transduction by dephosphorylating tyrosine residues on peptide substrates can induce apoptosis and halt cell multiplication.93 AgNPs can also alter the morphology of bacteria by obstructing the synthesis of cell wall peptidoglycan and inhibiting bacterial growth by disrupting the functions of the cell division protein FtsZ and the chromosomal replication initiator protein DnaA.94 Nevertheless, the potential mutagenic effects of AgNPs and the resistance mechanisms of target cells remain a subject of debate. Also, AgNPs have demonstrated the ability to disrupt biofilm formation by interfering with critical bacterial metabolic processes. They can hinder bacterial motility, impair iron uptake, induce oxidative stress, and disrupt respiration, which are crucial for bacterial survival.95 Additionally, AgNPs can interfere with quorum sensing systems, which bacteria use to communicate and coordinate biofilm development.96 Their small size also allows them to penetrate the exopolysaccharide layer via water channels, establishing direct contact with bacterial cells and inhibiting biofilm progression. This interaction leads to a reduction in membrane components, including proteins, polysaccharides, lipids, and nucleic acids, gradually compromising membrane stability.97 AgNPs exhibit potent antibacterial activity against a wide spectrum of bacterial species through multiple mechanisms. Fig. 3 illustrates key modes of action by which AgNPs combat bacterial pathogens, including disruption of cellular processes, membrane damage, and interference with biofilm formation.2.4. Resistance mechanisms of AgNPs
Silver-resistant bacteria were initially identified following their emergence from a burn wound subjected to silver nitrate treatment.98,99 Since then, these bacteria have been consistently detected in various environments, including clinical settings like hospitals and non-clinical environments such as mines and coastal waters.100,101 The mechanism of silver resistance was best described in S. typhimurium carrying the multidrug-resistant plasmid pMG101.99 This plasmid contains the sil operon, which plays a vital role in exogenous silver resistance due to horizontal gene transfer. The sil operon encodes a sophisticated resistance mechanism involving multiple genes, notably silCFBA, silE, and silRS. The SilCBA complex forms an efflux pump that facilitates the transfer of silver ions out of the bacterial cell, while SilE acts as a periplasmic silver-binding protein, and SilRS functions as a two-component regulatory system.102–104 The homolog sequences to the sil determinants have also been identified in numerous E. coli strains. The cusCFBA gene cluster, a component of the resistance-nodulation-division (RND) efflux system, is pivotal in the resistance mechanism against copper and silver.105 Lok et al. demonstrated that disrupting the cus locus in the silver-resistant strain led to a substantial reduction in the MIC of Ag+ from over 1000 μM to 12 μM.106 Concerning the outer membrane, it has been documented that E. coli mutants lacking outer membrane porins exhibit more resistance to Ag+ ions.107,108 The resistance mechanisms can also involve a phenotypic change rather than a genetic alteration. Panáček et al. found that Gram-negative bacteria can generate flagellin, an adhesive protein found in flagella, which induces the aggregation of AgNPs.109 The study proposed that suppressing flagellin production using pomegranate rind extract holds promise for overcoming resistance to AgNPs. Therefore, non-motile strains may exhibit increased susceptibility to AgNPs as the resistance mechanism is influenced by bacterial motility, particularly by the presence of the flagellum.110 Numerous studies have documented the response of bacterial biofilms to silver-induced stress.111 Gram-negative bacteria, particularly members of the Enterobacteriaceae family, appear more prone to developing silver resistance. Bacterial species exhibiting resistance to silver include Enterobacter spp.,112Pseudomonas spp.,113E. coli,114,115Klebsiella spp.,116Acinetobacter spp.,117Citrobacter spp.,118Morganella spp.,119Proteus spp.,120 and Cupriavidus spp.,121 among others. More recently, silver resistance has also been reported in some strains of S. aureus.122 The prevalence and mechanisms of silver resistance can vary among different strains and species.3. Vancomycin hydrochloride
Vancomycin, an amphoteric glycopeptide antibiotic, is frequently designated as the “drug of last resort”, typically administered after other antibiotics have failed.123,124 During the 1950s, researchers at Eli Lilly and Company, including Dr E.C. Kornfeld, isolated vancomycin (compound 05865) from Bornean soil samples. Produced by Streptomyces orientalis (now Amycolatopsis orientalis), it proved effective against Gram-positive bacteria, including antibiotic-resistant S. aureus. Its name, derived from “vanquish”, reflects its potency against resistant strains.125,126 Approved by the FDA in 1958, vancomycin's early use was limited by impurities and toxicity concerns, including ototoxicity and nephrotoxicity.127 Vancomycin is proven effective against S. aureus (MRSA), S. epidermidis, C. difficile, and it also has activity against other bacterial species, including Streptococcus spp., Enterococcus spp., L. monocytogenes, Actinomyces spp., Lactobacillus spp., and diphtheroids.128,129 Nonetheless, following the emergence of vancomycin-resistant enterococci strains in the late 1980s, the incidence of resistance in both enterococci and staphylococci has continued to rise.1303.1. Mechanism of antibacterial activity of vancomycin
Each class of antibiotics targets specific components or processes essential for bacterial survival and growth. They disrupt these processes through various mechanisms, including inhibiting cell wall and protein biosynthesis, disrupting cell membranes, and interfering with nucleic acid metabolism.131 The reported antibacterial mechanism for vancomycin is based on inhibiting the cross-linking of peptidoglycan chains, a vital structural element within the cell walls of bacteria.132 Peptidoglycan is a polysaccharide consisting of sugars and amino acids, with repeating units of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) in its sugar backbone crosslinked by oligopeptides attached to the NAM subunits.133 It is reported that vancomycin forms hydrogen bonds with the D-alanyl-D-alanine terminal (D-Ala-D-Ala) in the nascent peptidoglycan, thereby disrupting the synthesis of the bacterial cell wall.134,135 Due to steric hindrance, this binding inhibits the activity of enzymes such as transpeptidases which are crucial for cell wall synthesis.136 The lack of transpeptidation directly reduces the crosslinking density in the peptidoglycan matrix, resulting in decreased strength and stiffness in the cell, impairing the resistance to osmotic pressure, and leading to bacterial lysis.137,138In vitro evidence indicates that vancomycin also inhibits transglycosylation via binding to D-Ala-D-Ala moieties in peptidoglycan precursors such as lipid II, impeding the elongation of the peptidoglycan backbone.139,140 Besides inhibiting peptidoglycan biosynthesis, vancomycin has been found to affect genes involved in autolysis and peptidoglycan hydrolase (SagB), accelerating cell death.141 In addition, there are reports focusing on the effect of vancomycin on bacterial cell membranes, leading to alterations in membrane permeability and RNA synthesis.1283.2. Resistance mechanisms of vancomycin
Despite its effectiveness against Gram-positive bacteria, improper administration and excessive use of vancomycin can induce genetic alterations such as mutations or the acquisition of resistance genes. This culminates in the emergence of vancomycin-resistant strains like VRE, vancomycin-resistant S. aureus (VRSA), and vancomycin-intermediate S. aureus (VISA), characterized by higher MICs than non-mutated strains.142 It has been found that vancomycin resistance is directly attributed to the expression of gene clusters named van operons. Several established van operons (such as vanA, vanB, and vanD) encode enzymes that alter the biosynthesis of peptidoglycan precursors.143 This alteration produces substituted terminated precursors with less binding affinity to vancomycin, such as D-alanyl-D-lactate (D-Ala-D-Lac) and D-alanine-D-serine (D-Ala-D-Ser).144 One representative van operon, vanA, is shown in Fig. 4. It is found in both VRE and VRSA and contributes to the formation of D-Ala-D-Lac residue for transpeptidation.145,146 Upon activation, three enzymes, VanH, VanA, and VanX, are expressed from the vanA operon, which possess respective functions. The VanH reductase catalyzes the reduction of pyruvate to D-Lac, while the VanA ligase catalyzes the formation of a crucial bond between D-Lac and D-Ala through a covalent linkage.147 The VanX dipeptidase hydrolyzes the peptide bond in D-Ala-D-Ala residue.148 Additionally, the expression of the vanA cassette is controlled by a two-component regulatory system, VanR–VanS.149,150 It is also reported that the structure of peptide residue correlates with the level of vancomycin resistance, in which D-Ala-D-Lac rather than D-Ala-D-Ser is present in highly resistant genotypes of VRE, as the former structure shows less binding affinity.151,152 Besides van operons, there are also other reported genes associated with vancomycin resistance, such as cls and mprf, expressing enzymes to prevent vancomycin penetration via structural modifications of bacterial cell membranes.153–1554. Combination approaches for vancomycin and silver nanoparticles
Several studies have reported enhanced bactericidal effects when the combination of vancomycin and AgNPs is administered, surpassing the efficacy observed with either agent alone. This enhanced activity is valuable in overcoming bacterial antibiotic resistance mechanisms, thereby rejuvenating the effectiveness of vancomycin against resistant strains. The susceptibility of S. aureus biofilms to AgNPs and vancomycin was examined individually and in combination. Hair et al. used ten distinct S. aureus isolates, each representing a range of biofilm-forming abilities, and employed a crystal violet assay to quantify the mass of the biofilms.156 Treatment with 2 μg mL−1 vancomycin reduced the biofilm formation in 7 out of 10 isolates, encompassing 4 out of 5 methicillin-susceptible and 3 out of 5 methicillin-resistant S. aureus isolates. The AgNPs treatment demonstrated a decrease in 6 out of 10 isolates, including 4 out of 5 MSSA and 2 out of 5 MRSA strains, whereas the combinatorial treatment had a substantial reduction in biofilm formation in 9 out of 10 isolates. In another study, the combination of AgNPs with vancomycin demonstrated enhanced efficacy in inhibiting MRSA biofilms, showcasing a synergistic approach to combat antibiotic-resistant pathogens.157 AgNPs and vancomycin demonstrated promising synergistic antibiofilm activity with AgNP concentrations ranging from 2–4 μg mL−1, permitting non-cytotoxic application of the combination.4.1. Vancomycin conjugated to AgNPs
AgNP surface can be easily modified to increase its stability while offering an opportunity for drug conjugation. Thus, vancomycin can be conjugated to AgNPs based on the type of functional groups. Direct functionalization of AgNPs with vancomycin demonstrates the potential for developing novel antibacterial agents with enhanced efficacy. Esmaeillou et al. utilized N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) to conjugate vancomycin to the terminal carboxy group of thioglycolic acid-stabilized AgNPs.21 The resulting conjugate showed potent antibacterial activity, with MIC of 0.1 μg mL−1 against VRE and 0.05 μg mL−1 against S. aureus. Another study explored the use of polydopamine (pDA) coated AgNPs as a platform for vancomycin conjugation.158 It was found that the size of pDa–AgNPs formulation increased from 24 nm to 33 nm with vancomycin concentration in the micromolar range, potentially enhancing its efficacy against vancomycin-resistant E. faecalis and inhibiting further resistance development. Wang et al. took a different approach, designing vancomycin-modified magnetic-based silver microflowers assembly with bactericidal ability.159 The microflowers were carboxy-capped and then conjugated with vancomycin. The hybrid micro composite exhibited synergistic effects of magnetic-based Ag microflower and vancomycin for sterilization. The magnetic microflowers were designed to offer magnetic responsiveness, featuring a flower-like Ag shell to facilitate more release of Ag+ ions and bacterial contact, complemented by a vancomycin layer to enhance cell membrane permeability. The assembly demonstrated recyclability over five washing-and-reuse cycles and efficiently eradicated both E. coli and MRSA at low concentrations. In another study, poly(amidoamine) (PAMAM) dendrimers, conjugated with vancomycin via amide bonds and incorporating AgNPs, resulted in a 6–7 log reduction in colony-forming units (CFU) of a vancomycin-resistant bacterial strain in vitro.160 This treatment did not induce resistance in vancomycin-susceptible strains. Moreover, in vivo, bacterial killing was demonstrated in an infected wound murine model with a low dose of Van–PAMAM–AgNPs dendrimers upon its single topical application.A multifunctional platform capable of simultaneously detecting, eliminating, and inactivating pathogenic bacteria was constructed by Yang and coworkers.161 The electrochemical platform was fabricated using vancomycin-functionalized AgNPs/3D-ZnO nanorod array electrodes. AgNPs were functionalized with vancomycin using mercaptoacetic acid as a linker. This allowed for specific recognition and detection through hydrogen bonds between the peptidoglycan in the bacteria cell wall and the carbonyl and amine groups of vancomycin. This approach achieved a low detection limit of 330 CFU mL−1 and demonstrated effective bacterial elimination, with 50% efficiency at low concentrations (1000–2000 CFU mL−1). The synergistic effect of the AgNPs and vancomycin provided high antibacterial activity (99.99%) against pathogenic bacteria. Building on this concept of multifunctionality, Zhou et al. developed a nanocomplex that integrates surface-enhanced Raman scattering (SERS) and antimicrobial photodynamic therapy (aPDT) for theranostic applications against vancomycin-resistant bacterial infections.162 The nanocomplex was comprised of silver-coated gold nanoparticles encapsulated in silica functionalized with vancomycin and a near-infrared photosensitizer, silicon 2,3-naphthalocyanine dihydroxide. The SERS-active core facilitated sensitive SERS imaging of VRE strains, while the integrated photosensitizer generated ROS upon near-infrared light irradiation, resulting in the effective photodynamic killing of VRE. In vitro experiments demonstrated that the nano complex, when used at nanomolar concentrations, can significantly reduce bacterial populations by 4–5 log units upon photodynamic therapy.162 This finding indicates the nano complex's potential as an alternative method for bacterial detection and elimination. In another study, Zhang et al. synthesized vancomycin functionalized TiO2/AgNPs using nanoparticle deposition and chemical crosslinking reactions.163 These nanoparticles demonstrated remarkable efficacy in degrading methylene blue under ultraviolet (UV) illumination. The photocatalytic inactivation of sulfate-reducing bacteria Desulfotomaculum spp. was investigated under UV irradiation, and prepared nanoparticle assembly was able to prevent bacterial cell growth under UV irradiation.
4.2. Vancomycin electrostatic binding to AgNPs
AgNPs synthesized using an ionic surfactant can achieve stability through an electrostatic stabilization mechanism, where the surfactant acts as a crucial intermediary between the drug and the AgNPs.164 Notably, utilizing citrate capped AgNPs alongside vancomycin has yielded promising results, revealing a synergistic effect.165 In that study, the combination exhibited enhanced efficacy compared to free vancomycin against Gram-positive (S. aureus) and Gram-negative (E. coli) bacterial species. Further investigations were carried out to explore the possible mechanism of interaction of vancomycin functionalized AgNPs complex with bacterial cell walls. Vancomycin inhibits the synthesis of bacterial cell walls by binding to the D-alanyl-D-alanine dipeptide within the peptidoglycan layer.166 However, when combined with AgNPs, vancomycin exhibits a synergistic antibacterial effect through a four-step mechanism: (i) vancomycin binding to AgNPs, (ii) AgNPs–vancomycin complex interacting with bacteria, (iii) the release of Ag+ ions or AgNPs, and (iv) AgNPs inducing toxicity by binding to bacterial DNA and proteins.165 Another common approach involves the use of polymers or non-ionic surfactants, which interact with AgNPs through a steric repulsion mechanism.167,168 For instance, polyvinylpyrrolidone (PVP), a non-ionic polymer surfactant, has been used with citrate to functionalize vancomycin to AgNPs.169 Interestingly, citrate–AgNPs demonstrated better drug loading than PVP–AgNPs. Ma et al. loaded vancomycin on pDa nanoparticles through π–π stacking, electrostatic, and hydrogen-bonding interactions.170 AgNO3 was subsequently reduced in situ and embedded by the action of pDa from the pDa–vancomycin hybrid in a stable and facile process. Due to the robust interaction between pDA and vancomycin, the nanohybrid demonstrated sustained drug release during extended incubation periods. The incorporation of pDA helped stabilize the AgNPs, preventing their aggregation and thus maintaining their antibacterial effectiveness. In vitro testing using a spread plate assay revealed the superior antibacterial effect of the nanohybrid. While vancomycin and pDA–vancomycin treatments reduced bacterial colony numbers to 80.7% and 54.4%, respectively, the pDA–vancomycin–Ag nanohybrid group showed almost no bacterial colonies. This synergistic antibacterial effect was quantified by a low Combination Index (CI) value of 0.328. Additionally, the nanohybrid achieved hyperthermia-assisted bacterial inactivation due to the photothermal pDa. In S. aureus-infected mice, all wounds decreased in size over time, with the combined antibiotic-Ag treatment showing the most effective wound healing and significant reduction in wound area percentage. The authors suggested a possible antibacterial synergistic mechanism that disrupts the bacterial membrane and ROS generation.170 In another study, Veriato et al. investigated the formation of a complex between vancomycin–cysteamine and AgNPs.171 The synthesized nano-antibiotic exhibited potent bacteriostatic activity against both S. aureus and E. faecalis, achieving a MIC of ≤1 μg mL−1. These findings indicate that the antibiotic coating on AgNPs enhances both bacteriostatic and bactericidal effects, suggesting this approach is a promising alternative for treating resistant strains.Combining silver with other metals, such as gold, has been explored to enhance its antibacterial effects. Lu et al. prepared Au/Ag nanoparticles functionalized with vancomycin using a one-pot method.172 They observed that the antibacterial activity increased with Ag content, specifically when the Ag–Au ratio remained below 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Additionally, their study revealed that vancomycin could effectively enhance the adhesion between bacteria and the nanoparticles, effectively doubling it. The composite application showed lower bacterial resistance, with MIC values as low as 30 nmol mL−1.172 Hur and Park utilized a one-step, one-pot process to functionalize Au and Ag nanoparticles with vancomycin.173 In this process, vancomycin served dual roles as both a reducing agent and a capping agent for the nanoparticles. The process employed a green synthesis route to prepare spherically shaped particles with an average diameter of around 12 nm. Their report indicated that vancomycin–AgNPs (MIC 45.3–90.6 μg mL−1) have more antibacterial potential (2.4–4.8-fold increase) than vancomycin–AuNPs (MIC 217.4 or >217.4 μg mL−1).173
1. Additionally, their study revealed that vancomycin could effectively enhance the adhesion between bacteria and the nanoparticles, effectively doubling it. The composite application showed lower bacterial resistance, with MIC values as low as 30 nmol mL−1.172 Hur and Park utilized a one-step, one-pot process to functionalize Au and Ag nanoparticles with vancomycin.173 In this process, vancomycin served dual roles as both a reducing agent and a capping agent for the nanoparticles. The process employed a green synthesis route to prepare spherically shaped particles with an average diameter of around 12 nm. Their report indicated that vancomycin–AgNPs (MIC 45.3–90.6 μg mL−1) have more antibacterial potential (2.4–4.8-fold increase) than vancomycin–AuNPs (MIC 217.4 or >217.4 μg mL−1).173
In addition, silica nanoparticles have been used as carriers to enhance antibacterial functions for the co-delivery of vancomycin and AgNPs.174 The silica's porous structure facilitates a substantial loading capacity, whereas its spiky nano topography encourages pathogen interactions that enhance surface adhesion and localized delivery. A dual solvent method was employed to prepare Ag–silica nanoparticles, resulting in the uniform distribution of AgNPs measuring 10 to 25 nm on the surface of silica particles (∼200 nm) with a hollow core of approximately 110 nm, exhibiting no evident aggregation. Following this, vancomycin was loaded onto this composite, as shown demonstrating sustained release over two days and a synergistic bactericidal effect against both E. coli and S. epidermidis. The enhanced antibacterial effect of co-delivered nano-Ag and vancomycin using silica nanoparticles was explored through their interaction with bacterial membranes. Nano-Ag-decorated silica nanoparticles caused significant bacterial damage, effectively disrupting the membranes of E. coli and S. epidermidis.174
Hashimoto et al. explored the impact of a hydroxyapatite coating infused with silver (Ag–HA) and vancomycin on the MRSA biofilm formation.175 The study demonstrated that the Ag–HA coating effectively inhibited the formation of MRSA biofilms and altered their architecture. The combination of Ag–HA coating and vancomycin synergistically reduced the formation of MRSA biofilms both in vitro and in vivo. Sun et al. utilized layered double hydroxides (LDHs) to capture and disinfect bacteria simultaneously.176 The AgNPs were synthesized on the LDH surface via the reduction of Ag+ ions using glucose, followed by vancomycin adsorption onto the LDH sheets. The LDH nanocomposite demonstrated enhanced antibacterial activity against both Gram-positive and Gram-negative bacteria. In another study, Huang et al. developed pH-responsive platelet membrane-camouflaged nanoparticles of silver metal–organic framework (Ag–MOF) and vancomycin.177 The Ag–MOF was synthesized with 2-methylimidazole as the ligand and silver nitrate as the ion source. The vancomycin encapsulation efficiency was 81%, while the loading efficiency reached 64.7%. The Ag–MOF loaded with vancomycin exhibited significant antibacterial activity against standard clinical isolates in vitro, surpassing the efficacy of free vancomycin. The platelet membrane facilitates adhesion to the S. aureus surface and targets MRSA infection sites. In a mouse-MRSA pneumonia model, the nanocomposite demonstrated a superior anti-infective effect compared to free vancomycin without apparent toxicity.177
Biomaterials play a crucial role in modern medicine; however, treating infections associated with these materials remains challenging due to the increased antibiotic resistance of bacterial biofilms. Varisco et al. developed a self-protective antimicrobial and biocompatible coating utilizing Ag+ ions and a derivate of vancomycin, termed intelligent pyridinate vancomycin (IPV).178 Recently, Collatusso, et al. created a novel biological membrane from bovine pericardium samples impregnated with AgNPs or vancomycin to mitigate the drawbacks associated with antimicrobial properties in existing scaffold materials.179 The membrane successfully decreased the S. aureus loads in murine infection model, while also demonstrating good biocompatibility and osteoconductivity, indicating its potential applications in various clinical settings, particularly as an antimicrobial coating for implants in orthopedic and spinal surgery. In another study, AgNPs and vancomycin-loaded collagen nanofibers were synthesized using the electrospinning method.180 The synthesized nanofibers demonstrated antimicrobial activity against E. coli and S. aureus, emphasizing their promise for biomedical applications. Table 2 summarizes the antibacterial activity of vancomycin and AgNPs when used in combination.
| Sr. no. | Formulation | Particle size (nm) | Bacterial strain | Antibacterial activity | Ref. |
|---|---|---|---|---|---|
| Abbreviations: silver nanoparticles (AgNPs), minimum inhibitory effect (MIC), zone of inhibition (ZOI), minimal bactericidal concentration (MBC), combination index (CI), methicillin-resistant S. aureus (MRSA), methicillin-Sensitive S. aureus (MSSA), methicillin-resistant S. epidermidis (MRSE), vancomycin-resistant enterococci (VRE). | |||||
| 1 | AgNPs and vancomycin | MRSA: M1, M6, M7, HA1, USA300 GA-92. MSSA: SA 29213, SA 12600, SA 4651, SA 25923, SA 6538 | Out of 10 isolates, 9 showed a mean reduction in biofilm mass of 20.8 ± 3.8% after a combination treatment of 2 μg mL−1 AgNPs and 2 μg mL−1 vancomycin for 24 h | 156 | |
| AgNPs | 10 | ||||
| 2 | AgNPs and vancomycin | 10 S. aureus isolates were recovered from 50 urine specimens (7/10 MRSA) | MICs of AgNPs against MRSA isolates range from 1–2 μg ml−1. The AgNPs/vancomycin combination, used at MIC/MIC and 2MIC/4MIC, was more effective when the concentration of vancomycin was above the subinhibitory level (≥MIC) | 157 | |
| AgNPs | 20 to 42 | ||||
| 3 | Vancomycin-capped with thioglycolic acid-stabilized AgNPs | S. aureus, E. faecalis (VRE) S. epidermidis (MRSE), P. aeruginosa, E. coli | MIC for Vancomycin-capped AgNPs was 0.1 μg ml−1 for VRE, ≤0.02 μg ml−1 for MRSE, and 0.05 μg ml−1 for S. aureus | 21 | |
| AgNPs | 16 to 25 | ||||
| 4 | Vancomycin grafted polydopamine (pDA) coated AgNPs | Vancomycin-resistant E. faecalis | Vancomycin concentrations as low as 1–5 μM, when grafted to pDA–AgNPs, show a significant zone of inhibition and antibacterial efficacy compared to the standard high dose of vancomycin (13–54 mM) | 158 | |
| AgNPs | 12 ± 1.5 | ||||
| pDA–AgNPs | 24 ± 1.4 | ||||
| Vancomycin–pDA–AgNPs | 33 ± 1.6 | ||||
| 5 | Vancomycin-modified magnetic-based silver micro flowers (Van/Fe3O4–SiO2–Ag) | 700 to 900 | E. coli (BL21) and MRSA clinical isolates | MICs of the vancomycin-modified Fe3O4–SiO2–Ag microflowers were 10 μg mL−1 for E. coli and 20 μg mL−1 for MRSA | 159 |
| Fe3O4–SiO2–Ag micro flowers | |||||
| 6 | Poly-(amido-amine) (PAMAM) dendrimers with dual-conjugated vancomycin and AgNP (Van–PAMAM–AgNP) | S. aureus (ATCC12600) and S. aureus (Xen36) | Van–PAMAM–AgNP dendrimers achieved 6 to 7 log-unit reductions at 1 μg mL−1 Ag and 2 μg mL−1 vancomycin concentrations, primarily through cell wall damage. In mice with superficial wounds infected by vancomycin-resistant S. aureus Xen36, irrigation with low-dose Van–PAMAM–AgNP significantly improved healing compared to vancomycin or PAMAM–AgNP | 160 | |
| PAMAM–AgNP | 38 | ||||
| Van–PAMAM–AgNP dendrimers | 68 | ||||
| 7 | Vancomycin functionalized AgNPs/3D-ZnO nanorod array | — | E. coli and S. aureus | Platform demonstrated high detection sensitivity for S. aureus with a low detection limit of 330 CFU mL−1 and adaptable bacterial elimination efficiency (50%) at concentrations of 1000–2000 CFU mL−1. Additionally, it exhibited high antibacterial activity (99.99%) due to the synergistic germicidal effect of vancomycin and AgNPs | 161 |
| 8 | Silica-encapsulated silver-coated gold nanoparticles conjugated with 2,3-naphthalocyanine dihydroxide (Nc) and vancomycin (Au–AgNP–SiO2–Nc–Van) | B. subtilis (ATCC 6633), E. faecium (ATCC 51559, VanA), E. faecalis (ATCC 51299, VanB), and E. coli (ATCC 53868) | Au–AgNP–SiO2–Nc–Van showed a bacterial reduction of 99.999% upon irradiation with 54 J cm−2. Fast bacterial regression was observed during in vivo photodynamic therapy of mice with E. faecalis (VanB) infected wounds | 162 | |
| Au–AgNP–SiO2–Nc–Van | 110 | ||||
| Au–AgNPs | 60 | ||||
| 9 | Vancomycin functionalized silver–titanium oxide nanoparticles (Van–Ag–TiO2) | Sulphate-reducing bacteria (Desulfotomaculum spp.) | Vancomycin-sensitive sulfate-reducing bacteria were nearly eradicated within 1 h using Van–Ag–TiO2 under UV light irradiation, demonstrating high selective phototoxicity | 163 | |
| Van–Ag–TiO2 | 90 | ||||
| 10 | Vancomycin loaded citrate–AgNPs (Van–citrate–AgNPs) | S. aureus and E. coli | The zone of inhibition increased from 16 mm to 26.5 mm for S. aureus, while for E. coli, it increased from 0 mm to 7.5 mm | 165 | |
| AgNPs | 70 ± 3.3 | ||||
| Van–citrate–AgNPs 0.1 mM | 86 ± 4.5 | ||||
| Van–citrate–AgNPs 0.3 mM | 91 ± 2.3 | ||||
| 11 | Vancomycin–cysteamine complexed AgNPs (Van–Cys–AgNPs) | S. aureus (ATCC 29213) and the clinical strains (SA-1, SA-2, and SA-3), E. faecalis (ATCC 29212) and the clinical strains (EF-1, EF-2, and EF-3) | For S. aureus ATCC 2913, both the MIC (2.00 to 0.25 μg mL−1) and MBC (8 to 1 μg mL−1) exhibited an 8-fold reduction with Van–Cys–AgNPs treatment. For E. faecalis ATCC 29212, the MIC demonstrated a 16-fold reduction (2.0 to 0.12 μg mL−1) | 171 | |
| AgNPs | 1.51 to 43.82 | ||||
| 12 | Vancomycin-encapsulated polydopamine with AgNPs (pDA–Van–Ag) | E. coli and S. aureus | Vancomycin and pDA–Van treatments reduced colony numbers by 80.7% and 54.4%, respectively, whereas nearly no bacterial colonies were observed in the pDA–Van–Ag nanohybrid group for S. aureus. The combination index (CI) value was 0.328, indicating synergistic effects. Significant bacteria-killing activity was also observed in vivo using an S. aureus-infected mouse skin defect model | 170 | |
| pDA–Van–Ag | 290 | ||||
| 13 | Vancomycin-hybrid bimetallic nanoparticles (Au/AgNPs–Van) | E. coli ATCC 8099 and S. aureus ATCC 6538 | MICs against S. aureus were reduced to 30–60 nmol mL−1 for Au/AgNPs–Van-1 and AgNPs–Van, compared to 120 nmol mL−1 for vancomycin and AuNP–Van | 172 | |
| Ag/AuNPs–Van | 5 to 9 | ||||
| 14 | Vancomycin functionalized Au and Ag nanoparticles (Van–AuNPs) and (Van–AgNPs) | 19 MRSA strains and 1 MSSA strain (ATCC 29213) | MIC values for Van–AgNPs (45.3–90.6 μg mL−1) were lower than those of the Van–AuNPs (217.4 or >217.4 μg mL−1). Van–AgNPs demonstrated 2.4 to 4.8-fold more significant activity compared to Van–AuNPs | 173 | |
| Van–AuNPs | 11.01 ± 3.62 | ||||
| Van–AgNPs | 12.08 ± 2.13 | ||||
| 15 | Silica nanoparticles loaded with vancomycin and nano-silver (SiNPs–Ag–Van) | E. coli and S. epidermidis | MICs for E. coli and S. epidermidis were 15 and 10 μg mL−1, respectively, with the co-delivery system (SiNPs–Ag–Van) outperforming other formulations against both bacterial strains | 174 | |
| AgNPs | 10–25 | ||||
| Silica NPs | 200 | ||||
| 16 | Silver-containing hydroxyapatite (Ag–HA) coating and vancomycin | — | Clinical MRSA isolates | In vivo studies with Sprague-Dawley rats showed that Ag–HA combined with vancomycin was the most effective treatment compared to HA, HA–vancomycin, and Ag–HA | 175 |
| 17 | AgNPs and vancomycin co-modified layered double hydroxides (Van–Ag/LDHs) | E. coli and S. aureus | Van–Ag/LDHs platform demonstrated efficient antibacterial activity against E. coli and S. aureus, enabling simultaneous bacterial capture and disinfection | 176 | |
| AgNPs | 30 | ||||
| 18 | Platelet membrane-camouflaged vancomycin-loaded Ag–metal organic frameworks (PLT/Ag–MOF–Van) | E. coli (ATCC 25922), P. aeruginosa (ATCC 27853), and S. aureus (ATCC 25923) | MIC of free vancomycin was 2 μg mL−1, and Ag–MOF–Van was 1 μg mL−1 against MRSA, which was reduced to 0.5 μg mL−1 for PLT/Ag–MOF–Van. Encapsulation in the platelet membrane allowed it to bind effectively to MRSA bacteria and infection sites. PLT/Ag–MOF–Van showed significantly better anti-infective efficacy in vivo mouse MRSA pneumonia model compared to free vancomycin | 177 | |
| PLT/Ag–MOF–Van | 148 | ||||
| 19 | Vancomycin-derived molecule, intelligent pyridinate vancomycin (IPV) and silver | — | S. aureus 113, S. epidermidis 1457, and E. coli | MBC values for vancomycin and IPV were quasi-identical, highlighting the potential for use as biocompatible and self-protective antibacterial implant coatings | 178 |
| 20 | Bovine pericardium impregnated with vancomycin or AgNPs | 10 clinical isolates of MSSA and MRSA, each | Antibacterial effect were observed in an in vivo S. aureus infection mouse model following subcutaneous spine implantation, with minimal inflammatory response | 179 | |
| AgNPs | 50 | ||||
AgNPs have been shown to enhance the efficacy of vancomycin through various mechanisms. This integrated system offers a powerful strategy to overcome antibiotic resistance, as highlighted in multiple studies. Specifically, AgNPs have been demonstrated to significantly reduce the MIC of vancomycin by increasing oxidative stress, disrupting bacterial membranes, and facilitating antibiotic uptake, thereby improving its effectiveness against resistant bacterial strains. Morones-Ramirez et al. demonstrated that AgNPs play an essential role in restoring the bactericidal action of vancomycin against Gram-negative bacteria. The mechanism involved AgNPs increasing bacterial membrane permeability, allowing vancomycin to penetrate and become effective against Gram-negative pathogens. The combination therapy demonstrated significant reductions in bacterial cell counts. In vivo studies using mild and acute peritonitis models revealed that the combination of Ag+ and vancomycin significantly reduced E. coli cell counts, while individual treatments had limited effect.181 These findings suggest that combining AgNPs with vancomycin is a promising strategy to combat antibiotic-resistant infections and potentially repurpose existing antibiotics for broader application.
While combining AgNPs and vancomycin shows promise in enhancing antibacterial activity, there are several potential challenges to consider. The intricate synthesis process required to create a stable and effective AgNP vancomycin system poses technical hurdles, while concerns regarding the potential toxicity of these hybrid structures to human cells and tissues remain at the forefront of safety considerations. Moreover, the variable efficacy observed across different bacterial strains underscores the need for comprehensive testing and optimization to ensure broad-spectrum applicability. Long-term bacterial resistance development should also be considered. Regulatory approval and clinical translation present additional hurdles due to the novel nature of this nanomedicine approach. Addressing these challenges are essential to harness the full potential of combining AgNPs and vancomycin in combating antibiotic-resistant bacteria.
4.3. Other metal nanoparticles in enhancing antibiotic efficacy
The role of nanoparticles in combating bacterial resistance extends beyond silver, encompassing various metal nanoparticles such as copper, gold, and zinc. These nanoparticles, when combined with antibiotics, offer promising avenues for addressing antimicrobial resistance. Copper nanoparticles, for example, have shown strong antimicrobial activity against biofilm-forming pathogens,182 while gold nanoparticles are known for their ease of surface modification.183 These properties position other metallic nanoparticles as an attractive option for enhancing antibiotic delivery, similar to AgNPs. Table 3 presents some of the combinations of metallic nanoparticles with vancomycin.| Metal | Formulation | Size (nm) | Bacterial strain | Antibacterial activity | Ref. |
|---|---|---|---|---|---|
| Silver | Vancomycin–cysteamine complexed AgNPs (Van–Cys–AgNPs) | 1.51 to 43.82 | S. aureus (ATCC 29213) and the clinical strains (SA-1, SA-2, and SA-3), E. faecalis (ATCC 29212) and the clinical strains (EF-1, EF-2, and EF-3) | For S. aureus ATCC 2913, both the MIC (2.00 to 0.25) and MBC (8 to 1) exhibited an 8-fold reduction with Van–Cys–AgNPs treatment. For E. faecalis ATCC 29212, the MIC demonstrated a 16-fold reduction (2.0 to 0.12) | 171 |
| AgNPs | |||||
| Gold | Vancomycin-functionalized gold nanoparticles (V-GNPs) | 24 to 77 | E. coli (ATCC 25923), K. oxytoca (ATCC 43165), P. aeruginosa (NCIM 2036), S. aureus (ATCC 14222) | The observed antibacterial activities of V-GNPs were 1.4-, 1.6-, 1.8-, and 1.6-fold higher against E. coli, K. oxytoca, P. aeruginosa, and S. aureus, respectively, compared to pure vancomycin | 184 |
| Copper | Vancomycin-modified copper sulfide nanoparticles (CuS–Van) | 15 ± 5 | Van-resistant E. faecium (ATCC51559, VanA), E. faecalis (ATCC 51299, VanB) | CuS–Van with NIR irradiation demonstrated the highest antibacterial efficacy and the fastest infection regression in the in vivo study compared to the control groups | 185 |
| Iron | Vancomycin and nisin-modified magnetite (Fe3O4)–SiO2 nanostructures coated with chitosan | 22–47 | S. aureus ATCC 33591 (MRSA), S. aureus ATCC 25923 (MSSA) | Vancomycin-functionalized nanocomposites exhibited to be more efficient in eradicating bacterial cells both in vitro and in vivo | 186 |
| Gallium | Vancomycin and gallium nitrate (Ga (NO3)3) | — | MSSA (ATCC 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923) and MRSA (ATCC 33 923) and MRSA (ATCC 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 591) 591) |
The combination of vancomycin and Ga (NO3)3 reduced the MIC of vancomycin in MRSA, exhibiting an additive effect while also inhibiting biofilm formation and enhancing biofilm destruction, particularly in the MRSA strain | 187 |
| Nickel | Vancomycin-modified gold nanoparticles combined with magnetic nickel oxide nanoparticles (NiO NPs–AuNPs–Van) | 193.08 ± 1.61 | MRSA (800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 926) 926) |
Combination therapy accelerated wound healing in MRSA-infected mice by promoting collagen coverage, reducing IL-6 and TNF-α cytokines, and upregulating VEGF expression | 188 |
| Zinc | Vancomycin and zinc oxide nanoparticles | 20–150 | S. aureus clinical isolates | Decrease in vancomycin MICs from 2500–5000 μg mL−1 to 39–78.125 μg mL−1 when mixed with ZnO NPs | 189 |
5. Conclusion and future outlook
By pairing specialized properties of metallic nanoparticles with antibiotics, this method offers a dynamic approach to counteracting the growing threat of bacterial resistance. Metallic nanoparticles, including silver, copper, and gold, possess intrinsic antimicrobial properties that can augment the effectiveness of antibiotics. These nanoparticles work through mechanisms such as disrupting bacterial cell membranes, interfering with essential cellular processes, and inducing oxidative stress. This review highlights the synergistic potential of AgNPs combined with vancomycin as an effective approach to combat antibiotic-resistant pathogens. Insights gained from this combination can be applied more broadly to other antibiotics, enhancing their efficacy against resistant bacteria, particularly those residing in biofilms. Future research should focus on unraveling the molecular mechanisms behind how AgNPs boost antibiotic activity in resistant strains. Understanding the interactions between AgNPs and bacterial membranes, enzymes, and genetic pathways responsible for resistance will provide critical insights into optimizing these combinations. Additionally, studies should explore strategies to fine-tune nanoparticle size, surface charge, and coating materials to improve targeted drug delivery, minimize cytotoxicity, and ensure compatibility with specific antibiotics. From a clinical standpoint, comprehensive preclinical and clinical investigations are essential to assess the safety, efficacy, and optimal dosing of AgNPs in antibiotic formulations for both systemic and topical applications. Addressing concerns such as nanoparticle toxicity, long-term tissue accumulation, and environmental impact will be pivotal for securing regulatory approval and ensuring successful clinical translation. Collaborative efforts among researchers and clinicians are important to ensuring the stability, scalability, and cost-effectiveness of these technologies for widespread clinical adoption.In summary, ongoing research and optimization of metallic nanoparticle-antibiotic combinations have the potential to transform the treatment of antibiotic-resistant infections. By providing novel, effective therapeutic options, these strategies have the potential to improve treatment outcomes, alleviate healthcare burdens, and significantly enhance patient care in the face of the growing threat of antimicrobial resistance.
Abbreviations
| AgNPs | Silver nanoparticles |
| aPDT | Antimicrobial photodynamic therapy |
| CDC | Disease control and prevention |
| CFU | Colony-forming units |
| CI | Combination index |
| EDC | N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride |
| ETC | Electron transport chain |
| FDA | Food and Drug Administration |
| GLASS | Global antimicrobial resistance and use surveillance system |
| IPV | Intelligent pyridinate vancomycin |
| LDHs | Layered double hydroxides |
| MDR | Multidrug-resistant |
| MICs | Minimum inhibitory concentrations |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MRSE | Methicillin-resistant Staphylococcus epidermidis |
| NAM | N-Acetylmuramic acid |
| NAG | N-Acetylglucosamine |
| NHS | N-Hydroxysuccinimide |
| OECD | Organization for Economic Cooperation and Development |
| PAMAM | Poly(amidoamine) |
| PVP | Polypyrrolidine |
| RND | Resistance-nodulation-division |
| ROS | Reactive oxygen species |
| SEM | Scanning electron microscopy |
| SERS | Surface-enhanced Raman scattering |
| TCA | Tricarboxylic acid cycle |
| UV | Ultraviolet |
| VISA | vancomycin-intermediate S. aureus |
| VRE | Vancomycin-resistant enterococci |
| VRSA | Vancomycin-resistant S. aureus |
| WHO | World Health Organization |
Author contributions
Sakshi V. Khairnar: conceptualization, literature review, writing original draft & editing; Ashish Das: conceptualization, writing & editing, figure preparation (design and layout); David Oupický: editing, supervision; Marat Sadykov: editing, supervision, validation, investigation; Svetlana Romanova: conceptualization, editing, supervision, validation.Data availability
This review does not involve the analysis of primary research data, software, or code. The data tables presented in this study are compiled from publicly available sources in open-access journals cited throughout the manuscript. Full references to these sources are provided in the text. Additional data, supporting materials, or further clarifications will be made available upon reasonable request to the corresponding author.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The authors thank Dr Siwei Zhao and Dr Yang Wu for their constructive comments and guidance during the preparation of this manuscript.References
- Antimicrobial Resistance Collaborators, Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis, Lancet, 2022, 399(10325), 629–655, DOI:10.1016/S0140-6736(21)02724-0.
- J. M. Munita and C. A. Arias, Mechanisms of Antibiotic Resistance, Microbiol. Spectrum, 2016, 4(2), 481–511, DOI:10.1128/microbiolspec.VMBF-0016-2015.
- CDC, 2019 Antibiotic Resistance Threats Report. Antimicrobial Resistance, https://www.cdc.gov/antimicrobial-resistance/data-research/threats/index.html (accessed 2024-05-23).
- M. Naghavi, S. E. Vollset, K. S. Ikuta, L. R. Swetschinski, A. P. Gray, E. E. Wool, G. R. Aguilar, T. Mestrovic, G. Smith, C. Han, R. L. Hsu, J. Chalek, D. T. Araki, E. Chung, C. Raggi, A. G. Hayoon, N. D. Weaver, P. A. Lindstedt, A. E. Smith, U. Altay, N. V. Bhattacharjee, K. Giannakis, F. Fell, B. McManigal, N. Ekapirat, J. A. Mendes, T. Runghien, O. Srimokla, A. Abdelkader, S. Abd-Elsalam, R. G. Aboagye, H. Abolhassani, H. Abualruz, U. Abubakar, H. J. Abukhadijah, S. Aburuz, A. Abu-Zaid, S. Achalapong, I. Y. Addo, V. Adekanmbi, T. E. Adeyeoluwa, Q. E. S. Adnani, L. A. Adzigbli, M. S. Afzal, S. Afzal, A. Agodi, A. J. Ahlstrom, A. Ahmad, S. Ahmad, T. Ahmad, A. Ahmadi, A. Ahmed, H. Ahmed, I. Ahmed, M. Ahmed, S. Ahmed, S. A. Ahmed, M. A. Akkaif, S. A. Awaidy, Y. A. Thaher, S. O. Alalalmeh, M. T. AlBataineh, W. A. Aldhaleei, A. A. S. Al-Gheethi, N. B. Alhaji, A. Ali, L. Ali, S. S. Ali, W. Ali, K. Allel, S. Al-Marwani, A. Alrawashdeh, A. Altaf, A. B. Al-Tammemi, J. A. Al-Tawfiq, K. H. Alzoubi, W. A. Al-Zyoud, B. Amos, J. H. Amuasi, R. Ancuceanu, J. R. Andrews, A. Anil, I. A. Anuoluwa, S. Anvari, A. E. Anyasodor, G. L. C. Apostol, J. Arabloo, M. Arafat, A. Y. Aravkin, D. Areda, A. Aremu, A. A. Artamonov, E. A. Ashley, M. O. Asika, S. S. Athari, M. M. W. Atout, T. Awoke, S. Azadnajafabad, J. M. Azam, S. Aziz, A. Y. Azzam, M. Babaei, F.-X. Babin, M. Badar, A. A. Baig, M. Bajcetic, S. Baker, M. Bardhan, H. J. Barqawi, Z. Basharat, A. Basiru, M. Bastard, S. Basu, N. S. Bayleyegn, M. A. Belete, O. O. Bello, A. Beloukas, J. A. Berkley, A. S. Bhagavathula, S. Bhaskar, S. S. Bhuyan, J. A. Bielicki, N. I. Briko, C. S. Brown, A. J. Browne, D. Buonsenso, Y. Bustanji, C. G. Carvalheiro, C. A. Castañeda-Orjuela, M. Cenderadewi, J. Chadwick, S. Chakraborty, R. M. Chandika, S. Chandy, V. Chansamouth, V. K. Chattu, A. A. Chaudhary, P. R. Ching, H. Chopra, F. R. Chowdhury, D.-T. Chu, M. Chutiyami, N. Cruz-Martins, A. G. D. Silva, O. Dadras, X. Dai, S. D. Darcho, S. Das, F. P. D. L. Hoz, D. M. Dekker, K. Dhama, D. Diaz, B. F. R. Dickson, S. G. Djorie, M. Dodangeh, S. Dohare, K. G. Dokova, O. P. Doshi, R. K. Dowou, H. L. Dsouza, S. J. Dunachie, A. M. Dziedzic, T. Eckmanns, A. Ed-Dra, A. Eftekharimehrabad, T. C. Ekundayo, I. E. Sayed, M. Elhadi, W. El-Huneidi, C. Elias, S. J. Ellis, R. Elsheikh, I. Elsohaby, C. Eltaha, B. Eshrati, M. Eslami, D. W. Eyre, A. O. Fadaka, A. F. Fagbamigbe, A. Fahim, A. Fakhri-Demeshghieh, F. O. Fasina, M. M. Fasina, A. Fatehizadeh, N. A. Feasey, A. Feizkhah, G. Fekadu, F. Fischer, I. Fitriana, K. M. Forrest, C. F. Rodrigues, J. E. Fuller, M. A. Gadanya, M. Gajdács, A. P. Gandhi, E. E. Garcia-Gallo, D. O. Garrett, R. K. Gautam, M. W. Gebregergis, M. Gebrehiwot, T. G. Gebremeskel, C. Geffers, L. Georgalis, R. M. Ghazy, M. Golechha, D. Golinelli, M. Gordon, S. Gulati, R. D. Gupta, S. Gupta, V. K. Gupta, A. D. Habteyohannes, S. Haller, H. Harapan, M. L. Harrison, A. I. Hasaballah, I. Hasan, R. S. Hasan, H. Hasani, A. H. Haselbeck, M. S. Hasnain, I. I. Hassan, S. Hassan, M. S. H. Z. Tabatabaei, K. Hayat, J. He, O. E. Hegazi, M. Heidari, K. Hezam, R. Holla, M. Holm, H. Hopkins, M. M. Hossain, M. Hosseinzadeh, S. Hostiuc, N. R. Hussein, L. D. Huy, E. D. Ibáñez-Prada, A. Ikiroma, I. M. Ilic, S. M. S. Islam, F. Ismail, N. E. Ismail, C. D. Iwu, C. J. Iwu-Jaja, A. Jafarzadeh, F. Jaiteh, R. J. Yengejeh, R. D. G. Jamora, J. Javidnia, T. Jawaid, A. W. J. Jenney, H. J. Jeon, M. Jokar, N. Jomehzadeh, T. Joo, N. Joseph, Z. Kamal, K. K. Kanmodi, R. S. Kantar, J. A. Kapisi, I. M. Karaye, Y. S. Khader, H. Khajuria, N. Khalid, F. Khamesipour, A. Khan, M. J. Khan, M. T. Khan, V. Khanal, F. F. Khidri, J. Khubchandani, S. Khusuwan, M. S. Kim, A. Kisa, V. A. Korshunov, F. Krapp, R. Krumkamp, M. Kuddus, M. Kulimbet, D. Kumar, E. A. P. Kumaran, A. Kuttikkattu, H. H. Kyu, I. Landires, B. K. Lawal, T. T. T. Le, I. M. Lederer, M. Lee, S. W. Lee, A. Lepape, T. L. Lerango, V. S. Ligade, C. Lim, S. S. Lim, L. W. Limenh, C. Liu, X. Liu, X. Liu, M. J. Loftus, H. I. M. Amin, K. L. Maass, S. B. Maharaj, M. A. Mahmoud, P. Maikanti-Charalampous, O. M. Makram, K. Malhotra, A. A. Malik, G. D. Mandilara, F. Marks, B. A. Martinez-Guerra, M. Martorell, H. Masoumi-Asl, A. G. Mathioudakis, J. May, T. A. McHugh, J. Meiring, H. N. Meles, A. Melese, E. B. Melese, G. Minervini, N. S. Mohamed, S. Mohammed, S. Mohan, A. H. Mokdad, L. Monasta, A. M. Ghalibaf, C. E. Moore, Y. Moradi, E. Mossialos, V. Mougin, G. D. Mukoro, F. Mulita, B. Muller-Pebody, E. Murillo-Zamora, S. Musa, P. Musicha, L. A. Musila, S. Muthupandian, A. J. Nagarajan, P. Naghavi, F. Nainu, T. S. Nair, H. H. R. Najmuldeen, Z. S. Natto, J. Nauman, B. P. Nayak, G. T. Nchanji, P. Ndishimye, I. Negoi, R. I. Negoi, S. A. Nejadghaderi, Q. P. Nguyen, E. A. Noman, D. C. Nwakanma, S. O’Brien, T. J. Ochoa, I. A. Odetokun, O. A. Ogundijo, T. R. Ojo-Akosile, S. R. Okeke, O. C. Okonji, A. T. Olagunju, A. Olivas-Martinez, A. A. Olorukooba, P. Olwoch, K. I. Onyedibe, E. Ortiz-Brizuela, O. Osuolale, P. Ounchanum, O. T. Oyeyemi, P. A. Mahesh Padukudru, R. R. Parikh, J. Patel, S. Patil, S. Pawar, A. Y. Peleg, P. Peprah, J. Perdigão, C. Perrone, I.-R. Petcu, K. Phommasone, Z. Z. Piracha, D. Poddighe, A. J. Pollard, R. Poluru, A. Ponce-De-Leon, J. Puvvula, F. N. Qamar, N. H. Qasim, C. D. Rafai, P. Raghav, L. Rahbarnia, F. Rahim, V. Rahimi-Movaghar, M. Rahman, M. A. Rahman, H. Ramadan, S. K. Ramasamy, P. S. Ramesh, P. W. Ramteke, R. K. Rana, U. Rani, M.-M. Rashidi, D. Rathish, S. Rattanavong, S. Rawaf, E. M. M. Redwan, L. F. Reyes, T. Roberts, J. V. Robotham, V. D. Rosenthal, A. G. Ross, N. Roy, K. E. Rudd, C. J. Sabet, B. A. Saddik, M. R. Saeb, U. Saeed, S. S. Moghaddam, W. Saengchan, M. Safaei, A. Saghazadeh, N. S. Sharif-Askari, A. Sahebkar, S. S. Sahoo, M. Sahu, M. Saki, N. Salam, Z. Saleem, M. A. Saleh, Y. L. Samodra, A. M. Samy, A. Saravanan, M. Satpathy, A. E. Schumacher, M. Sedighi, S. Seekaew, M. Shafie, P. A. Shah, S. Shahid, M. J. Shahwan, S. Shakoor, N. Shalev, M. A. Shamim, M. A. Shamshirgaran, A. Shamsi, A. Sharifan, R. P. Shastry, M. Shetty, A. Shittu, S. Shrestha, E. E. Siddig, T. Sideroglou, J. Sifuentes-Osornio, L. M. L. R. Silva, E. A. F. Simões, A. J. H. Simpson, A. Singh, S. Singh, R. Sinto, S. S. M. Soliman, S. Soraneh, N. Stoesser, T. Z. Stoeva, C. K. Swain, L. Szarpak, S. Tabatabai, T. Y. Sree Sudha, Z. M.-A. Taha, K.-K. Tan, N. Tasak, N. Y. Tat, A. Thaiprakong, P. Thangaraju, C. C. Tigoi, K. Tiwari, M. R. Tovani-Palone, T. H. Tran, M. Tumurkhuu, P. Turner, A. J. Udoakang, A. Udoh, N. Ullah, S. Ullah, A. G. Vaithinathan, M. Valenti, T. Vos, H. T. L. Vu, Y. Waheed, A. S. Walker, J. L. Walson, T. Wangrangsimakul, K. G. Weerakoon, H. F. L. Wertheim, P. C. M. Williams, A. A. Wolde, T. M. Wozniak, F. Wu, Z. Wu, M. K. K. Yadav, S. Yaghoubi, Z. S. Yahaya, A. Yarahmadi, S. Yezli, Y. E. Yismaw, D. K. Yon, C.-W. Yuan, H. Yusuf, F. Zakham, G. Zamagni, H. Zhang, Z.-J. Zhang, M. Zielińska, A. Zumla, S. H. H. Zyoud, S. H. Zyoud, S. I. Hay, A. Stergachis, B. Sartorius, B. S. Cooper, C. Dolecek and C. J. L. Murray, Global Burden of Bacterial Antimicrobial Resistance 1990–2021: A Systematic Analysis with Forecasts to 2050, Lancet, 2024, 404(10459), 1199–1226, DOI:10.1016/S0140-6736(24)01867-1.
- A.-P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hindler, G. Kahlmeter, B. Olsson-Liljequist, D. L. Paterson, L. B. Rice, J. Stelling, M. J. Struelens, A. Vatopoulos, J. T. Weber and D. L. Monnet, Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance, Clin. Microbiol. Infect., 2012, 18(3), 268–281, DOI:10.1111/j.1469-0691.2011.03570.x.
- CDC. Antimicrobial Resistance Threats in the United States, 2021–2022. Antimicrobial Resistance. https://www.cdc.gov/antimicrobial-resistance/data-research/threats/update-2022.html (accessed 2024-07-21).
- WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed 2024-05-23).
- D. M. P. De Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Antimicrobial Resistance in ESKAPE Pathogens, Clin. Microbiol. Rev., 2020, 33(3), e00181-19, DOI:10.1128/CMR.00181-19.
- S. M. Mandal, A. Roy, A. K. Ghosh, T. K. Hazra, A. Basak and O. L. Franco, Challenges and Future Prospects of Antibiotic Therapy: From Peptides to Phages Utilization, Front. Pharmacol., 2014, 5, 105 Search PubMed.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, Metal Nanoparticles: Understanding the Mechanisms behind Antibacterial Activity, J. Nanobiotechnol., 2017, 15(1), 65, DOI:10.1186/s12951-017-0308-z.
- S. Tang and J. Zheng, Antibacterial Activity of Silver Nanoparticles: Structural Effects, Adv. Healthcare Mater., 2018, 7(13), e1701503, DOI:10.1002/adhm.201701503.
- F. Martinez-Gutierrez, L. Boegli, A. Agostinho, E. M. Sánchez, H. Bach, F. Ruiz and J. Garth, Anti-Biofilm Activity of Silver Nanoparticles against Different Microorganisms, Biofouling, 2013, 29(6), 651–660 CrossRef CAS PubMed.
- D. Davies, Understanding Biofilm Resistance to Antibacterial Agents, Nat. Rev. Drug Discovery, 2003, 2(2), 114–122, DOI:10.1038/nrd1008.
- H. Ceri, M. E. Olson, C. Stremick, R. R. Read, D. Morck and A. Buret, The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms, J. Clin. Microbiol., 1999, 37(6), 1771–1776, DOI:10.1128/jcm.37.6.1771-1776.1999.
- H. Deng, D. McShan, Y. Zhang, S. S. Sinha, Z. Arslan, P. C. Ray and H. Yu, Mechanistic Study of the Synergistic Antibacterial Activity of Combined Silver Nanoparticles and Common Antibiotics, Environ. Sci. Technol., 2016, 50(16), 8840–8848, DOI:10.1021/acs.est.6b00998.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles, Front. Microbiol., 2016, 7, 1831, DOI:10.3389/fmicb.2016.01831.
- K. Muppidi, J. Wang, G. Betageri and A. S. Pumerantz, PEGylated Liposome Encapsulation Increases the Lung Tissue Concentration of Vancomycin, Antimicrob. Agents Chemother., 2011, 55(10), 4537–4542, DOI:10.1128/AAC.00713-11.
- L. Bai, F. Lei, R. Luo, Q. Fei, Z. Zheng, N. He and S. Gui, Development of a Thermosensitive In situ Gel Formulations of Vancomycin Hydrochloride: Design, Preparation, In Vitro and In Vivo Evaluation, J. Pharm. Sci., 2022, 111(9), 2552–2561, DOI:10.1016/j.xphs.2022.04.011.
- M. Ansari, M. Shahlaei, S. Hosseinzadeh and S. Moradi, Recent Advances in Nanostructured Delivery Systems for Vancomycin, Nanomedicine, 2024, 19(23), 1931–1951, DOI:10.1080/17435889.2024.2377063.
- S. Pisani, S. Tufail, M. Rosalia, R. Dorati, I. Genta, E. Chiesa and B. Conti, Antibiotic-Loaded Nano-Sized Delivery Systems: An Insight into Gentamicin and Vancomycin, J. Funct. Biomater., 2024, 15(7), 194, DOI:10.3390/jfb15070194.
- M. Esmaeillou, G. Zarrini, M. Ahangarzadeh Rezaee, J. Shahbazi Mojarrad and A. Bahadori, Vancomycin Capped with Silver Nanoparticles as an Antibacterial Agent against Multi-Drug Resistance Bacteria, Adv. Pharm. Bull., 2017, 7(3), 479–483, DOI:10.15171/apb.2017.058.
- T. Akther, S. Priya, S. K. Sah, M. S. Khan and S. Hemalatha, Ta-AgNps Are Potential Antimicrobial Resistance Breakers, J. Nanostruct., 2019, 9(2), 376–383, DOI:10.22052/JNS.2019.02.019.
- K. Kon and M. Rai, Chapter 3 - Silver Nanoparticles for the Control of Vector-Borne Infections, in Nanotechnology in Diagnosis, Treatment and Prophylaxis of Infectious Diseases, ed. M. Rai and K. Kon, Academic Press, Boston, 2015, pp. 39–49. DOI:10.1016/B978-0-12-801317-5.00003-7.
- V. Edwards-Jones, The Benefits of Silver in Hygiene, Personal Care and Healthcare, Lett. Appl. Microbiol., 2009, 49(2), 147–152, DOI:10.1111/j.1472-765X.2009.02648.x.
- ACTISORB” Silver 220 Antimicrobial Binding Dressing. https://www.accessdata.fda.gov/cdrh_docs/pdf2/k022483.pdf (accessed 2024-05-25).
- Medical, A. Argentum Medical Announces 510(k) Clearance of Silverlon Burn and Wound Care Products for Vapor Sulfur Mustard Indication. https://www.silverlon.com/newsroom/argentum-medical-announces-510k-clearance-of-silverlon-burn-and-wound-care-products-for-vapor-sulfur-mustard-indication (accessed 2024-10-18).
- B. K. Pathi, S. Mishra, N. Moharana, A. Kanungo, A. Mishra, S. Sahu, R. K. Dash, R. Dubey and M. K. Das, Effect of Topical Silver Nanoparticle Formulation on Wound Bacteria Clearance and Healing in Patients With Infected Wounds Compared to Standard Topical Antibiotic Application: A Randomized Open-Label Parallel Clinical Trial, Cureus, 2024, 16(5), e60569, DOI:10.7759/cureus.60569.
- H. F. Hetta, I. M. S. Al-Kadmy, S. S. Khazaal, S. Abbas, A. Suhail, M. A. El-Mokhtar, N. H. A. Ellah, E. A. Ahmed, R. B. Abd-ellatief, E. A. El-Masry, G. E.-S. Batiha, A. A. Elkady, N. A. Mohamed and A. M. Algammal, Antibiofilm and Antivirulence Potential of Silver Nanoparticles against Multidrug-Resistant Acinetobacter Baumannii, Sci. Rep., 2021, 11(1), 10751, DOI:10.1038/s41598-021-90208-4.
- P. Siritongsuk, N. Hongsing and R. Patramanon, Two-Phase Bactericidal Mechanism of Silver Nanoparticles against Burkholderia Pseudomallei, PLoS One, 2016, 11(12), e0168098 CrossRef PubMed.
- J. M. Silvan, I. Zorraquin-Peña and A. J. Martinez-Rodriguez, Antibacterial Activity of Glutathione-Stabilized Silver Nanoparticles Against Campylobacter Multidrug-Resistant Strains, Frontiers, 2013, 458 Search PubMed.
- W.-R. Li, X.-B. Xie, Q.-S. Shi, H.-Y. Zeng, Y.-S. Ou-Yang and Y.-B. Chen, Antibacterial Activity and Mechanism of Silver Nanoparticles on Escherichia Coli, Appl. Microbiol. Biotechnol., 2010, 85(4), 1115–1122, DOI:10.1007/s00253-009-2159-5.
- A. H. Mondal, D. Yadav, A. Ali, N. Khan, J. O. Jin and Q. M. R. Haq, Anti-Bacterial and Anti-Candidal Activity of Silver Nanoparticles Biosynthesized Using Citrobacter Spp. MS5 Culture Supernatant, Biomolecules, 2020, 10(6), 944, DOI:10.3390/biom10060944.
- S. Gurunathan, J.-K. Jeong, J. W. Han, X.-F. Zhang, J. H. Park and J.-H. Kim, Multidimensional Effects of Biologically Synthesized Silver Nanoparticles in Helicobacter Pylori, Helicobacter Felis, and Human Lung (L132) and Lung Carcinoma A549 Cells, Nanoscale Res. Lett., 2015, 10(1), 35, DOI:10.1186/s11671-015-0747-0.
- M. H. Siddique, B. Aslam, M. Imran, A. Ashraf, H. Nadeem, S. Hayat, M. Khurshid, M. Afzal, I. R. Malik, M. Shahzad, U. Qureshi, Z. U. H. Khan and S. Muzammil, Effect of Silver Nanoparticles on Biofilm Formation and EPS Production of Multidrug-Resistant Klebsiella Pneumoniae, BioMed Res. Int., 2020, 2020, e6398165, DOI:10.1155/2020/6398165.
- L.-H. Li, M.-Y. Yen, C.-C. Ho, P. Wu, C.-C. Wang, P. K. Maurya, P.-S. Chen, W. Chen, W.-Y. Hsieh and H.-W. Chen, Non-Cytotoxic Nanomaterials Enhance Antimicrobial Activities of Cefmetazole against Multidrug-Resistant Neisseria Gonorrhoeae, PLoS One, 2013, 8(5), e64794, DOI:10.1371/journal.pone.0064794.
- A. J. Kora and J. Arunachalam, Assessment of Antibacterial Activity of Silver Nanoparticles on Pseudomonas Aeruginosa and Its Mechanism of Action, World J. Microbiol. Biotechnol., 2011, 27(5), 1209–1216, DOI:10.1007/s11274-010-0569-2.
- A. Parveen, M. S. Yalagatti, V. Abbaraju and R. Deshpande, Emphasized Mechanistic Antimicrobial Study of Biofunctionalized Silver Nanoparticles on Model Proteus Mirabilis, J. Drug Delivery, 2018, 2018, e3850139, DOI:10.1155/2018/3850139.
- A. K. Keshari, R. Srivastava, P. Singh, V. B. Yadav and G. Nath, Antioxidant and Antibacterial Activity of Silver Nanoparticles Synthesized by Cestrum Nocturnum, J. Ayurveda Integr. Med., 2020, 11(1), 37–44, DOI:10.1016/j.jaim.2017.11.003.
- S. Angamuthu, S. Thangaswamy, A. Raju, F. M. Husain, B. Ahmed, N. A. Al-Shabib, M. J. Hakeem, S. A. Shahzad, S. A. Abudujayn and S. Y. Alomar, Biogenic Preparation and Characterization of Silver Nanoparticles from Seed Kernel of Mangifera Indica and Their Antibacterial Potential against Shigella Spp, Molecules, 2023, 28(6), 2468, DOI:10.3390/molecules28062468.
- C. Krishnaraj, E. G. Jagan, S. Rajasekar, P. Selvakumar, P. T. Kalaichelvan and N. Mohan, Synthesis of Silver Nanoparticles Using Acalypha Indica Leaf Extracts and Its Antibacterial Activity against Water Borne Pathogens, Colloids Surf., B, 2010, 76(1), 50–56, DOI:10.1016/j.colsurfb.2009.10.008.
- F. K. Altuncu and A. Kamiloğlu, Optimization of Microwave-Assisted Biosynthesis of Silver Nanoparticle with Tarragon Extract, Biomass Convers. Biorefin., 2023, 13(6), 5419–5433, DOI:10.1007/s13399-022-02595-x.
- J. Li, K. Rong, H. Zhao, F. Li, Z. Lu and R. Chen, Highly Selective Antibacterial Activities of Silver Nanoparticles Against Bacillus Subtilis, J. Nanosci. Nanotechnol., 2013, 13(10), 6806–6813, DOI:10.1166/jnn.2013.7781.
- S. Suba, S. Vijayakumar, M. Nilavukkarasi, E. Vidhya and V. N. Punitha, Eco Synthesized Silver Nanoparticles as a next Generation of Nanoproduct in Multidisciplinary Applications, Environ. Chem. Ecotoxicol., 2022, 4, 13–19, DOI:10.1016/j.enceco.2021.11.001.
- O. Tverezovska, V. Holubnycha, R. Banasiuk, Y. Husak, S. Anton and V. Korniienko, The Effect of Silver Nanoparticles Against Formation of Enterococcus Faecalis Biofilms. In 2022 IEEE 12th International Conference Nanomaterials: Applications & Properties (NAP), 2022, pp. NRA12-1–NRA12-5. DOI:10.1109/NAP55339.2022.9934155.
- A. M. Grudniak, B. Milczarek, K. Markowska and K. I. Wolska, The Effect of Silver Nanoparticles on Listeria Monocytogenes PCM2191 Peptidoglycan Metabolism and Cell Permeability, 2018 Search PubMed.
- S. V. Patil, H. P. Borase, C. D. Patil and B. K. Salunke, Biosynthesis of Silver Nanoparticles Using Latex from Few Euphorbian Plants and Their Antimicrobial Potential, Appl. Biochem. Biotechnol., 2012, 167(4), 776–790, DOI:10.1007/s12010-012-9710-z.
- A. Banu and V. Rathod, Biosynthesis of Monodispersed Silver Nanoparticles and Their Activity against Mycobacterium Tuberculosis, Int. J. Biomed. Nanosci. Nanotechnol., 2013, 3(1–2), 211–220, DOI:10.1504/IJBNN.2013.054507.
- D. Swolana, M. Kępa, D. Idzik, A. Dziedzic, A. Kabała-Dzik, T. J. Wąsik and R. D. Wojtyczka, The Antibacterial Effect of Silver Nanoparticles on Staphylococcus Epidermidis Strains with Different Biofilm-Forming Ability, Nanomaterials, 2020, 10(5), 1010, DOI:10.3390/nano10051010.
- N. G. M. Attallah, E. Elekhnawy, W. A. Negm, I. A. Hussein, F. A. Mokhtar and O. M. Al-Fakhrany, In Vivo and In Vitro Antimicrobial Activity of Biogenic Silver Nanoparticles against Staphylococcus Aureus Clinical Isolates, Pharmaceuticals, 2022, 15(2), 194, DOI:10.3390/ph15020194.
- M. A. Pérez-Díaz, L. Boegli, G. James, C. Velasquillo, R. Sánchez-Sánchez, R.-E. Martínez-Martínez, G. A. Martínez-Castañón and F. Martinez-Gutierrez, Silver Nanoparticles with Antimicrobial Activities against Streptococcus Mutans and Their Cytotoxic Effect, Mater. Sci. Eng., C, 2015, 55, 360–366, DOI:10.1016/j.msec.2015.05.036.
- S. K. Mallineni, S. Sakhamuri, S. L. Kotha, A. R. G. M. AlAsmari, G. H. AlJefri, F. N. Almotawah, S. Mallineni and R. Sajja, Silver Nanoparticles in Dental Applications: A Descriptive Review, Bioengineering, 2023, 10(3), 327, DOI:10.3390/bioengineering10030327.
- G. Rehman, M. Umar, N. Shah, M. Hamayun, A. Ali, W. Khan, A. Khan, S. Ahmad, A. F. Alrefaei, M. H. Almutairi, Y.-S. Moon and S. Ali, Green Synthesis and Characterization of Silver Nanoparticles Using Azadirachta Indica Seeds Extract: In Vitro and In Vivo Evaluation of Anti-Diabetic Activity, Pharmaceuticals, 2023, 16(12), 1677, DOI:10.3390/ph16121677.
- H. Khajeh, B. Fazeli-Nasab, A. Pourshahdad, A. R. Mirzaei and M. Ghorbanpour, Green-Synthesized Silver Nanoparticles Induced Apoptotic Cell Death in CACO2 Cancer Cells by Activating MLH1 Gene Expression, Sci. Rep., 2024, 14(1), 29601, DOI:10.1038/s41598-024-80809-0.
- T. R. Sinclair, S. K. Hengel, B. G. van den Raza, S. A. Rutjes, A. M. D. R. Husman, W. J. G. M. Peijnenburg, H. E. D. W. Roesink and W. M. D. Vos, Surface Chemistry-Dependent Antiviral Activity of Silver Nanoparticles, Nanotechnology, 2021, 32(36), 365101, DOI:10.1088/1361-6528/ac03d6.
- M. R. Repon, T. Islam, H. T. Sadia, D. Mikučionienė, S. Hossain, G. Kibria and M. Kaseem, Development of Antimicrobial Cotton Fabric Impregnating AgNPs Utilizing Contemporary Practice, Coatings, 2021, 11(11), 1413, DOI:10.3390/coatings11111413.
- S. Kale, Emerging Agriculture Applications of Silver Nanoparticles, ES Food Agrofor., 2021, 17–22 CAS.
- E. O. Simbine, L. D. C. Rodrigues, J. Lapa-Guimarães, E. S. Kamimura, C. H. Corassin and C. A. F. D. Oliveira, Application of Silver Nanoparticles in Food Packages: A Review, Food Sci. Technol., 2019, 39, 793–802, DOI:10.1590/fst.36318.
- A. Sarhan, T. Fahmy and A. Habib, Optical, AC Conductivity and Antibacterial Activity Enhancement of Chitosan-Silver Nanocomposites for Optoelectronic and Biomedical Applications, Phys. Scr., 2024, 99(10), 105905, DOI:10.1088/1402-4896/ad7072.
- The Use of Silver Nanoparticles in Environmental Remediation[v1] | Preprints.org. https://www.preprints.org/manuscript/202301.0330/v1 (accessed 2025-02-06).
- M. Noga, J. Milan, A. Frydrych and K. Jurowski, Toxicological Aspects, Safety Assessment, and Green Toxicology of Silver Nanoparticles (AgNPs)—Critical Review: State of the Art, Int. J. Mol. Sci., 2023, 24(6), 5133, DOI:10.3390/ijms24065133.
- R. Bibi, I. A. Khan, I. Ullah, I. H. Khan, S. Razzaq, I. Ahmad, M. Fatima, F. U. Rehman, A. Ali, F. Khan and A. Riaz, Silver Nanoparticles: A Revolutionary Approach in Complementary and Alternative Medicine, Int. J. Vet. Sci., 2024, 2, 11–20, DOI:10.47278/book.CAM/2024.009.
- A. Karnwal, A. Y. Jassim, A. A. Mohammed, V. Sharma, A. R. M. S. Al-Tawaha and I. Sivanesan, Nanotechnology for Healthcare: Plant-Derived Nanoparticles in Disease Treatment and Regenerative Medicine, Pharmaceuticals, 2024, 17(12), 1711, DOI:10.3390/ph17121711.
- I. Khan, K. Saeed and I. Khan, Nanoparticles: Properties, Applications and Toxicities, Arabian J. Chem., 2019, 12(7), 908–931, DOI:10.1016/j.arabjc.2017.05.011.
- N. S. R. Rosman, N. A. Harun, I. Idris and W. I. Wan Ismail, Nanobiotechnology: Nature-Inspired Silver Nanoparticles towards Green Synthesis, Energy Environ., 2021, 32(7), 1183–1206, DOI:10.1177/0958305X21989883.
- P. Nie, Y. Zhao and H. Xu, Synthesis, Applications, Toxicity and Toxicity Mechanisms of Silver Nanoparticles: A Review, Ecotoxicol. Environ. Saf., 2023, 253, 114636, DOI:10.1016/j.ecoenv.2023.114636.
- N. P. U. Nguyen, N. T. Dang, L. Doan and T. T. H. Nguyen, Synthesis of Silver Nanoparticles: From Conventional to ‘Modern’ Methods—A Review, Processes, 2023, 11(9), 2617, DOI:10.3390/pr11092617.
- T. Mustapha, N. Misni, N. R. Ithnin, A. M. Daskum and N. Z. Unyah, A Review on Plants and Microorganisms Mediated Synthesis of Silver Nanoparticles, Role of Plants Metabolites and Applications, Int. J. Environ. Res. Public Health, 2022, 19(2), 674, DOI:10.3390/ijerph19020674.
- C. V. Restrepo and C. C. Villa, Synthesis of Silver Nanoparticles, Influence of Capping Agents, and Dependence on Size and Shape: A Review, Environ. Nanotechnol., Monit. Manage., 2021, 15, 100428, DOI:10.1016/j.enmm.2021.100428.
- A. K. Sidhu, N. Verma and P. Kaushal, Role of Biogenic Capping Agents in the Synthesis of Metallic Nanoparticles and Evaluation of Their Therapeutic Potential, Front. Nanotechnol., 2022, 3 DOI:10.3389/fnano.2021.801620.
- A. Naganthran, G. Verasoundarapandian, F. E. Khalid, M. J. Masarudin, A. Zulkharnain, N. M. Nawawi, M. Karim, C. A. C. Abdullah and S. A. Ahmad, Synthesis, Characterization and Biomedical Application of Silver Nanoparticles, Materials, 2022, 15(2), 427, DOI:10.3390/ma15020427.
- M. Tariq, K. N. Mohammad, B. Ahmed, M. A. Siddiqui and J. Lee, Biological Synthesis of Silver Nanoparticles and Prospects in Plant Disease Management, Molecules, 2022, 27(15), 4754, DOI:10.3390/molecules27154754.
- C. Wang, Y. J. Kim, P. Singh, R. Mathiyalagan, Y. Jin and D. C. Yang, Green Synthesis of Silver Nanoparticles by Bacillus Methylotrophicus, and Their Antimicrobial Activity, Artif. Cells, Nanomed., Biotechnol., 2016, 44(4), 1127–1132, DOI:10.3109/21691401.2015.1011805.
- D. Wang, B. Xue, L. Wang, Y. Zhang, L. Liu and Y. Zhou, Fungus-Mediated Green Synthesis of Nano-Silver Using Aspergillus Sydowii and Its Antifungal/Antiproliferative Activities, Sci. Rep., 2021, 11(1), 10356, DOI:10.1038/s41598-021-89854-5.
- P. Velmurugan, M. Cho, S.-S. Lim, S.-K. Seo, H. Myung, K.-S. Bang, S. Sivakumar, K.-M. Cho and B.-T. Oh, Phytosynthesis of Silver Nanoparticles by Prunus Yedoensis Leaf Extract and Their Antimicrobial Activity, Mater. Lett., 2015, 138, 272–275, DOI:10.1016/j.matlet.2014.09.136.
- J. Palanisamy, V. S. Palanichamy, G. Vellaichamy, P. Perumal, J. Vinayagam, S. Gunalan, S. G. Prabhakaran, P. P. Thiraviam, F. Musthafa, A. K. Balaraman and S. Rathinasamy, A Comprehensive Review on the Green Synthesis of Silver Nanoparticles from Marine Sources, Naunyn-Schmiedeberg’s Arch. Pharmacol., 2024, 1–24, DOI:10.1007/s00210-024-03547-0.
- Y. Zaman, M. Z. Ishaque, S. Ajmal, M. Shahzad, A. B. Siddique, M. U. Hameed, H. Kanwal, R. J. Ramalingam, M. Selvaraj and G. Yasin, Tamed Synthesis of AgNPs for Photodegradation and Anti-Bacterial Activity: Effect of Size and Morphology, Inorg. Chem. Commun., 2023, 150, 110523, DOI:10.1016/j.inoche.2023.110523.
- S. Agnihotri, S. Mukherji and S. Mukherji, Size-Controlled Silver Nanoparticles Synthesized over the Range 5–100 Nm Using the Same Protocol and Their Antibacterial Efficacy, RSC Adv., 2014, 4(8), 3974–3983, 10.1039/C3RA44507K.
- J. Helmlinger, C. Sengstock, C. Groß-Heitfeld, C. Mayer, T. A. Schildhauer, M. Köller and M. Epple, Silver Nanoparticles with Different Size and Shape: Equal Cytotoxicity, but Different Antibacterial Effects, RSC Adv., 2016, 6(22), 18490–18501, 10.1039/C5RA27836H.
- M. Rycenga, C. M. Cobley, J. Zeng, W. Li, C. H. Moran, Q. Zhang, D. Qin and Y. Xia, Controlling the Synthesis and Assembly of Silver Nanostructures for Plasmonic Applications, Chem. Rev., 2011, 111(6), 3669–3712, DOI:10.1021/cr100275d.
- S. Pal, Y. K. Tak and J. M. Song, Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-negative Bacterium Escherichia Coli, Appl. Environ. Microbiol., 2007, 73(6), 1712–1720, DOI:10.1128/AEM.02218-06.
- L. R. Pokhrel, Z. L. Jacobs, D. Dikin and S. M. Akula, Five Nanometer Size Highly Positive Silver Nanoparticles Are Bactericidal Targeting Cell Wall and Adherent Fimbriae Expression, Sci. Rep., 2022, 12(1), 6729, DOI:10.1038/s41598-022-10778-9.
- K. W. Fan and A. M. Granville, Surface Property Modification of Silver Nanoparticles with Dopamine-Functionalized Poly(Pentafluorostyrene) via RAFT Polymerization, Polymers, 2016, 8(3), 81, DOI:10.3390/polym8030081.
- A. Menichetti, A. Mavridi-Printezi, D. Mordini and M. Montalti, Effect of Size, Shape and Surface Functionalization on the Antibacterial Activity of Silver Nanoparticles, J. Funct. Biomater., 2023, 14(5), 244, DOI:10.3390/jfb14050244.
- C. Ashajyothi and R. Kelmani Chandrakanth, A Pilot Toxicology Study of Biogenic Silver Nanoparticles: In Vivo by Intraperitoneal and Intravenous Infusion Routes in Rats, J. Exp. Nanosci., 2019, 14(1), 89–106, DOI:10.1080/17458080.2018.1502479.
- H. A. Makeen and M. Albratty, Sesamol Loaded Silver Nanoparticles Gel Engineered for Wound Healing via Topical Delivery: Optimization In Vitro and Ex Vivo Evaluation, Curr. Pharm. Des., 2024, 30(40), 3175–3189, DOI:10.2174/0113816128306956240801052553.
- Y. He, J. Chen, Z. Xu, J. Nie, F. Wang, C. Ma, C. Wang, L. Zhang and C. Lu, Silver Functionalized Chitosan Composite Hydrogel with Sustained Silver Release and Enhanced Antibacterial Properties Promotes Healing of Infected Wounds, Int. J. Biol. Macromol., 2025, 285, 138290, DOI:10.1016/j.ijbiomac.2024.138290.
- I. X. Yin, J. Zhang, I. S. Zhao, M. L. Mei, Q. Li and C. H. Chu, The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry, Int. J. Nanomed., 2020, 15, 2555–2562, DOI:10.2147/IJN.S246764.
- O. Sedighi, B. Bednarke, H. Sherriff and A. L. Doiron, Nanoparticle-Based Strategies for Managing Biofilm Infections in Wounds: A Comprehensive Review, ACS Omega, 2024, 9(26), 27853–27871, DOI:10.1021/acsomega.4c02343.
- P. Wardman and L. P. Candeias, Fenton Chemistry: An Introduction, Radiat. Res., 1996, 145(5), 523–531, DOI:10.2307/3579270.
- A. Grzelak, M. Wojewódzka, S. Meczynska-Wielgosz, M. Zuberek, D. Wojciechowska and M. Kruszewski, Crucial Role of Chelatable Iron in Silver Nanoparticles Induced DNA Damage and Cytotoxicity, Redox Biol., 2018, 15, 435–440, DOI:10.1016/j.redox.2018.01.006.
- Q. L. Feng, J. Wu, G. Q. Chen, F. Z. Cui, T. N. Kim and J. O. Kim, A Mechanistic Study of the Antibacterial Effect of Silver Ions on Escherichia Coli and Staphylococcus Aureus, J. Biomed. Mater. Res., 2000, 52(4), 662–668, DOI:10.1002/1097-4636(20001215)52:4<662::AID-JBM10>3.0.CO;2-3.
- M. A. Radzig, V. A. Nadtochenko, O. A. Koksharova, J. Kiwi, V. A. Lipasova and I. A. Khmel, Antibacterial Effects of Silver Nanoparticles on Gram-negative Bacteria: Influence on the Growth and Biofilms Formation, Mechanisms of Action, Colloids Surf., B, 2013, 102, 300–306, DOI:10.1016/j.colsurfb.2012.07.039.
- D. Bamal, A. Singh, G. Chaudhary, M. Kumar, M. Singh, N. Rani, P. Mundlia and A. R. Sehrawat, Silver Nanoparticles Biosynthesis, Characterization, Antimicrobial Activities, Applications, Cytotoxicity and Safety Issues: An Updated Review, Nanomaterials, 2021, 11(8), 2086, DOI:10.3390/nano11082086.
- B. Liu, D. Liu, T. Chen, X. Wang, H. Xiang, G. Wang and R. Cai, iTRAQ-Based Quantitative Proteomic Analysis of the Antibacterial Mechanism of Silver Nanoparticles against Multidrug-Resistant Streptococcus Suis, Front. Microbiol., 2023, 14, 1293363, DOI:10.3389/fmicb.2023.1293363.
- N. Tripathi and M. K. Goshisht, Recent Advances and Mechanistic Insights into Antibacterial Activity, Antibiofilm Activity, and Cytotoxicity of Silver Nanoparticles, ACS Appl. Bio Mater., 2022, 5(4), 1391–1463, DOI:10.1021/acsabm.2c00014.
- E. Paluch, J. Rewak-Soroczyńska, I. Jędrusik, E. Mazurkiewicz and K. Jermakow, Prevention of Biofilm Formation by Quorum Quenching, Appl. Microbiol. Biotechnol., 2020, 104(5), 1871–1881, DOI:10.1007/s00253-020-10349-w.
- N. Hussein and M. M. Khadum, Evaluation of the Biosynthesized Silver Nanoparticles” Effects on Biofilm Formation, J. Appl. Sci. Nanotechnol., 2021, 1(1), 23–31, DOI:10.53293/jasn.2021.11019.
- C. Jelenko, Silver Nitrate Resistant E. Coli: Report of Case, Ann. Surg., 1969, 170(2), 296–299 CrossRef PubMed.
- G. Larkin Mchugh, R. C. Moellering, C. C. Hopkins and M. N. Swartz, Salmonella Typhimurium Resistant to Silver Nitrate, Chloramphenicol, and Ampicillin: A New Threat in Burn Units?, Lancet, 1975, 305(7901), 235–240, DOI:10.1016/S0140-6736(75)91138-1.
- C. Haefeli, C. Franklin and K. Hardy, Plasmid-Determined Silver Resistance in Pseudomonas Stutzeri Isolated from a Silver Mine, J. Bacteriol., 1984, 158(1), 389–392, DOI:10.1128/jb.158.1.389-392.1984.
- P. Choudhury and R. Kumar, Multidrug- and Metal-Resistant Strains of Klebsiella Pneumoniae Isolated from Penaeus Monodon of the Coastal Waters of Deltaic Sundarban, Can. J. Microbiol., 1998, 44(2), 186–189, DOI:10.1139/w97-144.
- A. Gupta, K. Matsui, J.-F. Lo and S. Silver, Molecular Basis for Resistance to Silver Cations in Salmonella, Nat. Med., 1999, 5(2), 183–188, DOI:10.1038/5545.
- H. Wang, J. Li, C. Min, F. Xia, M. Tang, J. Li, Y. Hu and M. Zou, Characterization of Silver Resistance and Coexistence of sil Operon with Antibiotic Resistance Genes Among Gram-negative Pathogens Isolated from Wound Samples by Using Whole-Genome Sequencing, Infect. Drug Resist., 2022, 15, 1425–1437, DOI:10.2147/IDR.S358730.
- S. Franke, Microbiology of the Toxic Noble Metal Silver, in Molecular Microbiology of Heavy Metals, ed. D. H. Nies and S. Silver, Springer, Berlin, Heidelberg, 2007, pp. 343–355. DOI:10.1007/7171_2006_084.
- S. Franke, G. Grass, C. Rensing and D. H. Nies, Molecular Analysis of the Copper-Transporting Efflux System CusCFBA of Escherichia Coli, J. Bacteriol., 2003, 185(13), 3804–3812, DOI:10.1128/jb.185.13.3804-3812.2003.
- C.-N. Lok, C.-M. Ho, R. Chen, P. K.-H. Tam, J.-F. Chiu and C.-M. Che, Proteomic Identification of the Cus System as a Major Determinant of Constitutive Escherichia Coli Silver Resistance of Chromosomal Origin, J. Proteome Res., 2008, 7(6), 2351–2356, DOI:10.1021/pr700646b.
- A. P. Pugsley and C. A. Schnaitman, Outer Membrane Proteins of Escherichia Coli VII. Evidence That Bacteriophage-Directed Protein 2 Functions as a Pore, J. Bacteriol., 1978, 133(3), 1181–1189 CrossRef CAS PubMed.
- X. Z. Li, H. Nikaido and K. E. Williams, Silver-Resistant Mutants of Escherichia Coli Display Active Efflux of Ag+ and Are Deficient in Porins, J. Bacteriol., 1997, 179(19), 6127–6132, DOI:10.1128/jb.179.19.6127-6132.1997.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek, O. Tomanec and R. Zbořil, Bacterial Resistance to Silver Nanoparticles and How to Overcome It, Nat. Nanotechnol., 2018, 13(1), 65–71, DOI:10.1038/s41565-017-0013-y.
- L. M. Stabryla, K. A. Johnston, N. A. Diemler, V. S. Cooper, J. E. Millstone, S.-J. Haig and L. M. Gilbertson, Role of Bacterial Motility in Differential Resistance Mechanisms of Silver Nanoparticles and Silver Ions, Nat. Nanotechnol., 2021, 16(9), 996–1003, DOI:10.1038/s41565-021-00929-w.
- S. Sudheer Khan, E. Bharath Kumar, A. Mukherjee and N. Chandrasekaran, Bacterial Tolerance to Silver Nanoparticles (SNPs): Aeromonas Punctata Isolated from Sewage Environment, J. Basic Microbiol., 2011, 51(2), 183–190, DOI:10.1002/jobm.201000067.
- S. Sütterlin, M. Dahlö, C. Tellgren-Roth, W. Schaal and Å. Melhus, High Frequency of Silver Resistance Genes in Invasive Isolates of Enterobacter and Klebsiella Species, J. Hosp. Infect., 2017, 96(3), 256–261, DOI:10.1016/j.jhin.2017.04.017.
- N. Hachicho, P. Hoffmann, K. Ahlert and H. J. Heipieper, Effect of Silver Nanoparticles and Silver Ions on Growth and Adaptive Response Mechanisms of Pseudomonas Putida Mt-2, FEMS Microbiol. Lett., 2014, 355(1), 71–77, DOI:10.1111/1574-6968.12460.
- J. L. Graves, M. Tajkarimi, Q. Cunningham, A. Campbell, H. Nonga, S. H. Harrison and J. E. Barrick, Rapid Evolution of Silver Nanoparticle Resistance in Escherichia Coli, Front. Genet., 2015, 6 DOI:10.3389/fgene.2015.00042.
- M. Muller, Bacterial Silver Resistance Gained by Cooperative Interspecies Redox Behavior, Antimicrob. Agents Chemother., 2018, 62(8), e00672–e00618, DOI:10.1128/AAC.00672-18.
- A. T. Hendry and I. O. Stewart, Silver-Resistant Enterobacteriaceae from Hospital Patients, Can. J. Microbiol., 1979, 25(8), 915–921, DOI:10.1139/m79-136.
- L. M. Deshpande and B. A. Chopade, Plasmid Mediated Silver Resistance in Acinetobacter Baumannii, BioMetals, 1994, 7(1), 49–56, DOI:10.1007/BF00205194.
- J. T. Trevors, Silver Resistance and Accumulation in Bacteria, Enzyme Microb. Technol., 1987, 9(6), 331–333, DOI:10.1016/0141-0229(87)90054-8.
- R. Y. Parikh, S. Singh, B. L. V. Prasad, M. S. Patole, M. Sastry and Y. S. Shouche, Extracellular Synthesis of Crystalline Silver Nanoparticles and Molecular Evidence of Silver Resistance from Morganella Sp.: Towards Understanding Biochemical Synthesis Mechanism, ChemBioChem, 2008, 9(9), 1415–1422, DOI:10.1002/cbic.200700592.
- A. Abbaszadegan, Y. Ghahramani, A. Gholami, B. Hemmateenejad, S. Dorostkar, M. Nabavizadeh and H. Sharghi, The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram-positive and Gram-negative Bacteria: A Preliminary Study, J. Nanomater., 2015, 2015, e720654, DOI:10.1155/2015/720654.
- S. Monchy, M. A. Benotmane, P. Janssen, T. Vallaeys, S. Taghavi, D. van der Lelie and M. Mergeay, Plasmids pMOL28 and pMOL30 of Cupriavidus Metallidurans Are Specialized in the Maximal Viable Response to Heavy Metals, J. Bacteriol., 2007, 189(20), 7417–7425, DOI:10.1128/jb.00375-07.
- J. V. Loh, S. L. Percival, E. J. Woods, N. J. Williams and C. A. Cochrane, Silver Resistance in MRSA Isolated from Wound and Nasal Sources in Humans and Animals, Int. Wound J., 2009, 6(1), 32–38, DOI:10.1111/j.1742-481X.2008.00563.x.
- Y. Fu, Y. Xu, F. Ruijne and O. P. Kuipers, Engineering Lanthipeptides by Introducing a Large Variety of RiPP Modifications to Obtain New-to-Nature Bioactive Peptides, FEMS Microbiol. Rev., 2023, 47(3), fuad017, DOI:10.1093/femsre/fuad017.
- D. H. Williams, The Glycopeptide Story – How to Kill the Deadly ‘Superbugs’, Nat. Prod. Rep., 1996, 13(6), 469–477, 10.1039/NP9961300469.
- R. S. Griffith, Introduction to Vancomycin, Rev. Infect. Dis., 1981, 3(Supplement_2), S200–S204, DOI:10.1093/clinids/3.Supplement_2.S200.
- O. V. Kisil, T. A. Efimenko and O. V. Efremenkova, Looking Back to Amycolatopsis: History of the Antibiotic Discovery and Future Prospects, Antibiotics, 2021, 10(10), 1254, DOI:10.3390/antibiotics10101254.
- E. Rubinstein and Y. Keynan, Vancomycin Revisited – 60 Years Later, Front. Public Health, 2014, 2, 217, DOI:10.3389/fpubh.2014.00217.
- C. Watanakunakorn, The Antibacterial Action of Vancomycin, Rev. Infect. Dis., 1981, 3(suppl), S210–S215 CrossRef PubMed.
- F. Soriano, J. Zapardiel and E. Nieto, Antimicrobial Susceptibilities of Corynebacterium Species and Other Non-Spore-Forming Gram-positive Bacilli to 18 Antimicrobial Agents, Antimicrob. Agents Chemother., 1995, 39(1), 208–214, DOI:10.1128/aac.39.1.208.
- A. Fodor, B. A. Abate, P. Deák, L. Fodor, E. Gyenge, M. G. Klein, Z. Koncz, J. Muvevi, L. Ötvös, G. Székely, D. Vozik and L. Makrai, Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review, Pathogens, 2020, 9(7), 522, DOI:10.3390/pathogens9070522.
- S. Kırmusaoğlu, Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods, BoD – Books on Demand, 2019 Search PubMed.
- S. Patel, C. V. Preuss and F. Bernice, Vancomycin, in StatPearls, StatPearls Publishing, Treasure Island (FL), 2024 Search PubMed.
- B. P. Howden, J. K. Davies, P. D. R. Johnson, T. P. Stinear and M. L. Grayson, Reduced Vancomycin Susceptibility in Staphylococcus Aureus, Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications, Clin. Microbiol. Rev., 2010, 23(1), 99–139 CrossRef CAS PubMed.
- C. C. McComas, B. M. Crowley and D. L. Boger, Partitioning the Loss in Vancomycin Binding Affinity for D-Ala-d-Lac into Lost H-Bond and Repulsive Lone Pair Contributions, J. Am. Chem. Soc., 2003, 125(31), 9314–9315, DOI:10.1021/ja035901x.
- J. D. Buynak, Cutting and Stitching: The Cross-Linking of Peptidoglycan in the Assembly of the Bacterial Cell Wall, ACS Chem. Biol., 2007, 2(9), 602–605, DOI:10.1021/cb700182u.
- J. R. Knox and R. F. Pratt, Different Modes of Vancomycin and D-Alanyl-D-Alanine Peptidase Binding to Cell Wall Peptide and a Possible Role for the Vancomycin Resistance Protein, Antimicrob. Agents Chemother., 1990, 7, 1342–1347, DOI:10.1128/aac.34.7.1342.
- P. Loskill, P. M. Pereira, P. Jung, M. Bischoff, M. Herrmann, M. G. Pinho and K. Jacobs, Reduction of the Peptidoglycan Crosslinking Causes a Decrease in Stiffness of the Staphylococcus Aureus Cell Envelope, Biophys. J., 2014, 107(5), 1082–1089, DOI:10.1016/j.bpj.2014.07.029.
- H. Jiang and S. X. Sun, Morphology, Growth, and Size Limit of Bacterial Cells, Phys. Rev. Lett., 2010, 105(2), 028101, DOI:10.1103/PhysRevLett.105.028101.
- R. Sinha Roy, P. Yang, S. Kodali, Y. Xiong, R. M. Kim, P. R. Griffin, H. R. Onishi, J. Kohler, L. L. Silver and K. Chapman, Direct Interaction of a Vancomycin Derivative with Bacterial Enzymes Involved in Cell Wall Biosynthesis, Chem. Biol., 2001, 8(11), 1095–1106, DOI:10.1016/S1074-5521(01)00075-8.
- L. Cegelski, S. J. Kim, A. W. Hing, D. R. Studelska, R. D. O'Connor, A. K. Mehta and J. Schaefer, Rotational-Echo Double Resonance Characterization of the Effects of Vancomycin on Cell Wall Synthesis in Staphylococcus Aureus, Biochemistry, 2002, 41(43), 13053–13058, DOI:10.1021/bi0202326.
- B. Salamaga, L. Kong, S. J. Foster, L. Pasquina-Lemonche, L. Lafage, M. von und zur Muhlen, J. F. Gibson and D. Grybchuk, Demonstration of the Role of Cell Wall Homeostasis in Staphylococcus Aureus Growth and the Action of Bactericidal Antibiotics, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(44) DOI:10.1073/pnas.2106022118.
- R. Rengaraj, S. Mariappan, U. Sekar and A. Kamalanadhan, Detection of Vancomycin Resistance among Enterococcus Faecalis and Staphylococcus Aureus, J. Clin. Diagn. Res., 2016, 10(2), DC04–DC06, DOI:10.7860/JCDR/2016/17552.7201.
- M. O. Ahmed and K. E. Baptiste, Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health, Microb. Drug Resist., 2018, 24(5), 590–606, DOI:10.1089/mdr.2017.0147.
- G. Li, M. J. Walker and D. M. P. De Oliveira, Vancomycin, Resistance in Enterococcus and Staphylococcus Aureus, Microorganisms, 2023, 11(1), 24, DOI:10.3390/microorganisms11010024.
- C. M. Hill, K. M. Krause, S. R. Lewis, J. Blais, B. M. Benton, M. Mammen, P. P. Humphrey, A. Kinana and J. W. Janc, Specificity of Induction of the vanA and vanB Operons in Vancomycin-Resistant Enterococci by Telavancin, Antimicrob. Agents Chemother., 2010, 54(7), 2814–2818, DOI:10.1128/aac.01737-09.
- W. A. McGuinness, N. Malachowa and F. R. DeLeo, Vancomycin Resistance in Staphylococcus Aureus, Yale J. Biol. Med., 2017, 90(2), 269–281 CAS.
- M. Arthur, P. Reynolds and P. Courvalin, Glycopeptide Resistance in Enterococci, Trends Microbiol., 1996, 4(10), 401–407, DOI:10.1016/0966-842X(96)10063-9.
- I. A. D. Lessard and C. T. Walsh, VanX, a Bacterial d-Alanyl-d-Alanine Dipeptidase: Resistance, Immunity, or Survival Function?, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(20), 11028, DOI:10.1073/pnas.96.20.11028.
- A. A. Guffey and P. J. Loll, Regulation of Resistance in Vancomycin-Resistant Enterococci: The VanRS Two-Component System, Microorganisms, 2021, 9(10), 2026, DOI:10.3390/microorganisms9102026.
- M. Arthur, C. Molinas and P. Courvalin, The VanS-VanR Two-Component Regulatory System Controls Synthesis of Depsipeptide Peptidoglycan Precursors in Enterococcus Faecium BM4147, J. Bacteriol., 1992, 174(8), 2582–2591, DOI:10.1128/jb.174.8.2582-2591.1992.
- F. Lebreton, F. Depardieu, N. Bourdon, M. Fines-Guyon, P. Berger, S. Camiade, R. Leclercq, P. Courvalin and V. Cattoir, D-Ala-d-Ser VanN-Type Transferable Vancomycin Resistance in Enterococcus Faecium, Antimicrob. Agents Chemother., 2011, 55(10), 4606–4612, DOI:10.1128/aac.00714-11.
- P. J. Stogios and A. Savchenko, Molecular Mechanisms of Vancomycin Resistance, Protein Sci., 2020, 29(3), 654–669, DOI:10.1002/pro.3819.
- K. Thitiananpakorn, Y. Aiba, X.-E. Tan, S. Watanabe, K. Kiga, Y. Sato'o, T. Boonsiri, F.-Y. Li, T. Sasahara, Y. Taki, A. H. Azam, Y. Zhang and L. Cui, Association of mprF Mutations with Cross-Resistance to Daptomycin and Vancomycin in Methicillin-Resistant Staphylococcus Aureus (MRSA), Sci. Rep., 2020, 10(1), 16107, DOI:10.1038/s41598-020-73108-x.
- W. Li, J. Hu, L. Li, M. Zhang, Q. Cui, Y. Ma, H. Su, X. Zhang, H. Xu and M. Wang, New Mutations in Cls Lead to Daptomycin Resistance in a Clinical Vancomycin- and Daptomycin-Resistant Enterococcus Faecium Strain, Front. Microbiol., 2022, 13, 896916, DOI:10.3389/fmicb.2022.896916.
- A. M. Turner, J. Y. H. Lee, C. L. Gorrie, B. P. Howden and G. P. Carter, Genomic Insights Into Last-Line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-Resistant Enterococcus, Front. Microbiol., 2021, 12 DOI:10.3389/fmicb.2021.637656.
- B. B. Hair, M. E. Conley, T. M. Wienclaw, M. J. Conley and B. K. Berges, Synergistic Activity of Silver Nanoparticles and Vancomycin Against a Spectrum of Staphylococcus Aureus Biofilm Types, bioRxiv, 2018, 337436, DOI:10.1101/337436.
- D. Doaa Safwat Mohamed, Silver Nanoparticles -Vancomycin Combination: A Powerful Synergism with Promising Anti Biofilm Activity against High Virulence Mrsa Isolates, New Egypt. J. Microbiol., 2019, 53, 13–26 Search PubMed.
- K. Patel and R. Kumar, Vancomycin Grafted Polydopamine Coated Silver Nanoparticles for Enhanced Antibacterial Action Against Vancomycin–Resistant Bacteria E. Faecalis. - Patel - 2020 - ChemistrySelect - Wiley Online Library.
- C. Wang, K. Zhang, Z. Zhou, Q. Li, L. Shao, R. Z. Hao, R. Xiao and S. Wang, Vancomycin-Modified Fe3O4@SiO2@Ag Microflowers as Effective Antimicrobial Agents, Int. J. Nanomed., 2017, 12, 3077–3094, DOI:10.2147/IJN.S132570.
- G. Jiang, S. Liu, T. Yu, R. Wu, Y. Ren, H. C. van der Mei, J. Liu and H. J. Busscher, PAMAM Dendrimers with Dual-Conjugated Vancomycin and Ag-Nanoparticles Do Not Induce Bacterial Resistance and Kill Vancomycin-Resistant Staphylococci, Acta Biomater., 2021, 123, 230–243, DOI:10.1016/j.actbio.2021.01.032.
- Z. Yang, Y. Wang and D. Zhang, A Novel Multifunctional Electrochemical Platform for Simultaneous Detection, Elimination, and Inactivation of Pathogenic Bacteria Based on the Vancomycin-Functionalised AgNPs/3D-ZnO Nanorod Arrays, Biosens. Bioelectron., 2017, 98, 248–253, DOI:10.1016/j.bios.2017.06.058.
- Z. Zhou, S. Peng, M. Sui, S. Chen, L. Huang, H. Xu and T. Jiang, Multifunctional Nanocomplex for Surface-Enhanced Raman Scattering Imaging and near-Infrared Photodynamic Antimicrobial Therapy of Vancomycin-Resistant Bacteria, Colloids Surf., B, 2018, 161, 394–402, DOI:10.1016/j.colsurfb.2017.11.005.
- Y. Wan, D. Zhang, Y. Wang, P. Qi, J. Wu and B. Hou, Vancomycin-Functionalised Ag@TiO2 Phototoxicity for Bacteria, J. Hazard. Mater., 2011, 186(1), 306–312, DOI:10.1016/j.jhazmat.2010.10.110.
- H. Alsubaie, Z. Zaheer and E. S. Aazam, Role of Ionic Surfactants on the Nucleation and Growth of Silver Nanoparticles, J. Mol. Liq., 2021, 341, 117309, DOI:10.1016/j.molliq.2021.117309.
- A. Kaur, S. Preet, V. Kumar, R. Kumar and R. Kumar, Synergetic Effect of Vancomycin Loaded Silver Nanoparticles for Enhanced Antibacterial Activity, Colloids Surf., B, 2019, 176, 62–69, DOI:10.1016/j.colsurfb.2018.12.043.
- G. D. Wright and C. T. Walsh, D-Alanyl-D-Alanine Ligases and the Molecular Mechanism of Vancomycin Resistance, Acc. Chem. Res., 1992, 25(10), 468–473, DOI:10.1021/ar00022a006.
- L. M. Liz-Marzán and I. Lado-Touriño, Reduction and Stabilization of Silver Nanoparticles in Ethanol by Nonionic Surfactants, Langmuir, 1996, 12(15), 3585–3589, DOI:10.1021/la951501e.
- M. Tejamaya, I. Römer, R. C. Merrifield and J. R. Lead, Stability of Citrate, PVP, and PEG Coated Silver Nanoparticles in Ecotoxicology Media, Environ. Sci. Technol., 2012, 46(13), 7011–7017, DOI:10.1021/es2038596.
- A. Kaur, D. Goyal and R. Kumar, Surfactant Mediated Interaction of Vancomycin with Silver Nanoparticles, Appl. Surf. Sci., 2018, 449, 23–30, DOI:10.1016/j.apsusc.2017.12.066.
- K. Ma, P. Dong, M. Liang, S. Yu, Y. Chen and F. Wang, Facile Assembly of Multifunctional Antibacterial Nanoplatform Leveraging Synergistic Sensitization between Silver Nanostructure and Vancomycin, ACS Appl. Mater. Interfaces, 2020, 12(6), 6955–6965, DOI:10.1021/acsami.9b22043.
- T. S. Veriato, I. Fontoura and M. L. Castilho, Nano-Antibiotic Based on Silver Nanoparticles Functionalized to the Vancomycin–Cysteamine Complex for Treating Staphylococcus Aureus and Enterococcus Faecalis, Pharmacol. Rep., 2023, 75, 951–961 CrossRef CAS PubMed.
- Z. Lu, J. Zhang, Z. Yu, X. Liu, Z. Zhang, W. Wang, X. Wang, Y. Wang and D. Wang, Vancomycin-Hybrid Bimetallic Au/Ag Composite Nanoparticles: Preparation of the Nanoparticles and Characterization of the Antibacterial Activity, New J. Chem., 2017, 41(13), 5276–5279, 10.1039/C7NJ01660C.
- Y. E. Hur and Y. Park, Vancomycin-Functionalized Gold and Silver Nanoparticles as an Antibacterial Nanoplatform Against Methicillin-Resistant Staphylococcus Aureus, J. Nanosci. Nanotechnol., 2016, 16(6), 6393–6399 CrossRef CAS PubMed.
- C. Ni, Y. Zhong, W. Wu, Y. Song, P. Makvandi, C. Yu and H. Song, Co-Delivery of Nano-Silver and Vancomycin via Silica Nanopollens for Enhanced Antibacterial Functions, Antibiotics, 2022, 11(5), 685, DOI:10.3390/antibiotics11050685.
- A. Hashimoto, H. Miyamoto, T. Kobatake, T. Nakashima, T. Shobuike, M. Ueno, T. Murakami, I. Noda, M. Sonohata and M. Mawatari, The Combination of Silver-Containing Hydroxyapatite Coating and Vancomycin Has a Synergistic Antibacterial Effect on Methicillin-Resistant Staphylococcus Aureus Biofilm Formation, Bone Jt. Res., 2020, 9(5), 211–218, DOI:10.1302/2046-3758.95.BJR-2019-0326.R1.
- J. Sun, J. Li, H. Fan and S. Ai, Ag Nanoparticles and Vancomycin Comodified Layered Double Hydroxides for Simultaneous Capture and Disinfection of Bacteria, J. Mater. Chem. B, 2013, 1(40), 5436–5442, 10.1039/C3TB20871K.
- R. Huang, G.-Q. Cai and R. Gui, Platelet Membrane-Camouflaged Silver Metal-Organic Framework Drug System against Infections Caused by Methicillin-Resistant Staphylococcus Aureus, J. Nanobiotechnol., 2021, 19(229), 1–19 Search PubMed.
- M. Varisco, N. Khanna, P. S. Brunetto and K. M. Fromm, New Antimicrobial and Biocompatible Implant Coating with Synergic Silver–Vancomycin Conjugate Action, ChemMedChem, 2014, 9(6), 1221–1230, DOI:10.1002/cmdc.201400072.
- D. D. F. F. Collatusso, L. R. Dantas, P. H. Suss, J. L. Rocha, M. D. P. Loureiro and F. F. Tuon, Biological Membrane with Antimicrobial Activity Based on Lyophilized and Decellularized Bovine Pericardium Impregnated with Vancomycin or Silver Nanoparticle, 2023. DOI:10.20944/preprints202307.0646.v1.
- Antimicrobial electrospun collagen nanofibers loaded with Silver nanoparticles and Vancomycin. https://www.jstage.jst.go.jp/article/jpssuppl/WCP2018/0/WCP2018_PO4-11-17/_article/-char/ja/ (accessed 2024-05-26).
- J. R. Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Silver Enhances Antibiotic Activity Against Gram-negative Bacteria, Sci. Transl. Med., 2013, 5(190), 190ra81, DOI:10.1126/scitranslmed.3006276.
- F. LewisOscar, D. MubarakAli, C. Nithya, R. Priyanka, V. Gopinath, N. S. Alharbi and N. Thajuddin, One Pot Synthesis and Anti-Biofilm Potential of Copper Nanoparticles (CuNPs) against Clinical Strains of Pseudomonas Aeruginosa, Biofouling, 2015, 31(4), 379–391, DOI:10.1080/08927014.2015.1048686.
- E.-K. Tian, Y. Wang, R. Ren, W. Zheng and W. Liao, Gold Nanoparticle: Recent Progress on Its Antibacterial Applications and Mechanisms, J. Nanomater., 2021, 2021(1), 2501345, DOI:10.1155/2021/2501345.
- T. A. Hagbani, H. Yadav, A. Moin, A. S. A. Lila, K. Mehmood, F. Alshammari, S. Khan, E.-S. Khafagy, T. Hussain, S. M. D. Rizvi and M. H. Abdallah, Enhancement of Vancomycin Potential against Pathogenic Bacterial Strains via Gold Nano-Formulations: A Nano-Antibiotic Approach, Materials, 2022, 15(3), 1108, DOI:10.3390/ma15031108.
- Z. Zou, J. Sun, Q. Li, Y. Pu, J. Liu, R. Sun, L. Wang and T. Jiang, Vancomycin Modified Copper Sulfide Nanoparticles for Photokilling of Vancomycin-Resistant Enterococci Bacteria, Colloids Surf., B, 2020, 189, 110875, DOI:10.1016/j.colsurfb.2020.110875.
- M. Nasaj, A. Farmany, L. Shokoohizadeh, F. A. Jalilian, R. Mahjoub, G. Roshanaei, A. Nourian, O. H. Shayesteh and M. Arabestani, Vancomycin and Nisin-Modified Magnetic Fe3O4@SiO2 Nanostructures Coated with Chitosan to Enhance Antibacterial Efficiency against Methicillin Resistant Staphylococcus Aureus (MRSA) Infection in a Murine Superficial Wound Model, BMC Chem., 2024, 18(1), 43, DOI:10.1186/s13065-024-01129-y.
- N. Mohammad Hanifeh, S. Keyvani-Ghamsari, K. Khorsandi and E. Mahmoodi Khaledi, Effect of Gallium Nitrate on the Antibacterial Activity of Vancomycin in Methicillin-Sensitive and Resistant Staphylococcus Aureus, Arch. Microbiol., 2024, 206(7), 304, DOI:10.1007/s00203-024-04028-x.
- T. Du, J. Cao, Z. Xiao, J. Liu, L. Wei, C. Li, J. Jiao, Z. Song, J. Liu, X. Du and S. Wang, Van-Mediated Self-Aggregating Photothermal Agents Combined with Multifunctional Magnetic Nickel Oxide Nanoparticles for Precise Elimination of Bacterial Infections, J. Nanobiotechnol., 2022, 20(1), 325, DOI:10.1186/s12951-022-01535-1.
- R. Thamer and E. Alsammak, Synergistic Effect of Zinc Oxide Nanoparticles and Vancomycin on Methicillin Resistant Staphylococcus Aureus, J. Life Bio Sci. Res., 2021, 2(01), 01–06, DOI:10.38094/jlbsr1332.
| This journal is © The Royal Society of Chemistry 2025 |