Open Access Article
Open Access ArticleMeniran (Phyllanthus niruri L.) embedded zeolitic imidazolate framework (ZIF-8) nanoparticle for cancer chemotherapy: supported molecular docking analysis†
Fasih Bintang Ilhami *a,
Sari Edi Cahyaningrum
*a,
Sari Edi Cahyaningrum b,
Andika Pramudya Wardanab,
Noto Susanto Gultomc,
Hasan Subektia,
Astrid Rahmawatid and
Sapti Puspitarini*a
b,
Andika Pramudya Wardanab,
Noto Susanto Gultomc,
Hasan Subektia,
Astrid Rahmawatid and
Sapti Puspitarini*a
aDepartment of Natural Science Faculty of Mathematics and Natural Science, Universitas Negeri Surabaya, Surabaya 60231, Indonesia. E-mail: fasihilhami@unesa.ac.id; saptipuspitarini@unesa.ac.id
bDepartment of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Negeri Surabaya, Surabaya 60231, Indonesia
cDepartment of Physics, Universitas Padjadjaran, Bandung 45363, Indonesia
dDepartment of Applied Chemistry, Osaka Institute of Technology, Osaka 535-8585, Japan
First published on 2nd January 2025
Abstract
Cancer is among the leading causes of mortality worldwide. Natural bioactive compounds like Meniran (Phyllanthus niruri L.) have been the focus of extensive research due to their potent anticancer properties. Nevertheless, drug delivery strategies may be necessary to encapsulate bioactive compounds, thereby reducing their toxicity and enhancing their stability. Herein, we successfully synthesized Meniran extract incorporated zeolitic imidazolate framework (ZIF-8) nanoparticles for anticancer therapy. Meniran-incorporated ZIF-8 nanoparticles possess unique advantages including well-distributed nanoparticles with rhombic dodecahedrons and excellent pH-responsiveness. In vitro analysis showed that Meniran-incorporated ZIF-8 nanoparticles have anticancer activity towards HeLa cells. Interestingly, computational simulations offer valuable insights into the molecular-level interaction mechanisms between ZIF-8 and specific proteins under cancer cells. As far as we are aware, this is the first report of natural bioactive compounds derived from Meniran encapsulated into nanoparticles as a drug delivery system, marking a significant advancement in the development of novel biomaterials for cancer treatment.
Introduction
Conventional cancer treatments are widely utilized but suffer from several drawbacks, including inadequate, solubility of the drug, lack of specificity towards the target, and notable adverse effects. 1,2 Drug delivery system-based nanoparticles present promising alternatives as therapeutic agents, with the potential to substantially enhance cancer treatment efficacy. Drug-loaded nanoparticles possess the capability to pass through the vascular network of tumors owing to the Enhanced Permeability and Retention (EPR) effect. This phenomenon extends the amount of time that anticancer drugs stay in bloodstream, subsequently improving drug bioavailability and therapeutic efficacy while reducing the occurrence of side effects. 3–7 The EPR effect has been substantiated in multiple tumor models, wherein nanoparticle-drug conjugates achieve drug concentrations 10- to 100-fold higher than free drugs within tumor tissues. This targeted drug delivery mechanism could have been the driving force of the EPR effect exclusive to tumor tissues and was not observed in normal tissues. 8–10 The primary objective of cancer research has been the successful delivery of targeted drugs to specific areas of the body, to minimize adverse effects and maximize therapeutic potential.To date, the focus on plant-derived natural compounds has garnered attention due to their phytochemicals for therapeutics. Phyllanthus niruri L., referred to known as Meniran is a medicinal plant that commonly thrives in humid and rocky regions especially Indonesia. The plant comprises a diverse array of chemical components, including as lignans, tannins, polyphenols, alkaloids, flavonoids, terpenoids, and steroids.11 Nurcholis and coworkers12 mentioned that Meniran is notably rich in phenolic compounds. Most importantly, Meniran has been investigated as an anticancer agent in several cancer cell lines, such as Lewis cells line, 13,14 HL-60 cells line, 15 HepG2 cells line, 16 143B cell line, 17 and A549 cells line. 14,18 Thus, Meniran has pharmacological efficacy due to its bioactive components such as hepatoprotective, reducing blood sugar levels, anticancer, antimicrobial, preventing oxidative damage, and cardioprotective. 19 However, despite the notable advantages of plant-derived natural compounds, there are still various obstacles to overcome in terms of their biomedical application including their solubility and stability in water, as well as potential harmful effects within the body. 20 As a possible approach to tackle these issues, we hypothesized that incorporating plant-derived natural compounds into smart materials might have a significant alter on their solubility and stability in water.
Metal–Organic Frameworks (MOF) represent a continuously expanding crystalline materials classification consists metal ions and an organic component known as the linker. 21 MOF possess numerous attractive characteristics including their significant porosity (which exceeds 50% of the crystal volume), large surface area (ranging from about 1000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m2 g−1), and the ability to precisely adjust their properties by altering either the reaction conditions or the reactants. MOF have been applied in a diverse array of fields, including catalysis, gas storage, adsorbents for separation processes, sensors, biomedical imaging, and drug delivery. 22–27 Zeolitic Imidazolate Framework (ZIF-8) is a very notable metal–organic framework known for its remarkable properties, mostly attributed to its significant pore width of 11.6 Å and nanosized. These properties include facile synthesis at room temperature, economically viable precursors, excellent stability, high surface area, and the potential for further enhancements through chemical modification of the linker which promising candidate for drug delivery systems. 28–33 ZIF-8 has a unique characteristic of pH-responsive drug release, whereby pharmaceuticals are released at a faster rate at acidic pH levels (5.5–6), which align with the internal conditions of cancer cells, namely inside endosomes and lysosomes. As a consequence, this leads to more targeted drug release, thereby minimizing the side effects associated with the carried drugs. 28,34,35
000 m2 g−1), and the ability to precisely adjust their properties by altering either the reaction conditions or the reactants. MOF have been applied in a diverse array of fields, including catalysis, gas storage, adsorbents for separation processes, sensors, biomedical imaging, and drug delivery. 22–27 Zeolitic Imidazolate Framework (ZIF-8) is a very notable metal–organic framework known for its remarkable properties, mostly attributed to its significant pore width of 11.6 Å and nanosized. These properties include facile synthesis at room temperature, economically viable precursors, excellent stability, high surface area, and the potential for further enhancements through chemical modification of the linker which promising candidate for drug delivery systems. 28–33 ZIF-8 has a unique characteristic of pH-responsive drug release, whereby pharmaceuticals are released at a faster rate at acidic pH levels (5.5–6), which align with the internal conditions of cancer cells, namely inside endosomes and lysosomes. As a consequence, this leads to more targeted drug release, thereby minimizing the side effects associated with the carried drugs. 28,34,35
Herein, we create plant-derived natural compounds of extract Meniran incorporated into ZIF-8 nanoparticles for a potential approach to cancer therapy. These materials clearly showed well-distribution nanoparticles with strong activity towards cancer cells. Moreover, computational simulations provide insight into the interaction mechanisms of ZIF-8 with specific proteins at the molecular level. To the best of our knowledge, this is the first report on the incorporation of bioactive compounds from extract Meniran into ZIF-8 nanoparticles for cancer therapy. Therefore, this study could offer valuable insights into the development of innovative strategies based on plant-derived natural compound nanoparticles for cancer treatment Scheme 1.
 | ||
| Scheme 1 Graphical illustration Meniran-incorporated ZIF-8 nanoparticles and release within cancer cells to provide toxicity. | ||
Results and discussion
The Meniran extract was obtained through the extraction method, which employed methanol as the solvent, subsequently obtained through column separation following solvent evaporation. First, the LC-HRMS was conducted to confirm the qualitative results of compounds isolated within Meniran. As shown in Table S1†, the bioactive compound analysis presented that Meniran contains several bioactive compounds which include flavonoid, tannin, and phenol group. These results indicate that the combination extract contains bioactive compounds that have pharmacological effects as anticancer. The therapeutic and pharmacological effects of plant-derived natural chemicals are attributed to their active components, such as phenol and flavonoid groups, which have been scientifically shown to possess anticancer properties. 38–40 Next, we also performed interaction prediction between Meniran compounds and proteins by in silico study. Meniran extract is anticipated to potentially suppress cancer cell proliferation due to its active components, which include quercetin, rutin, and quercetin 3-beta-D-glucoside (Isoquercitrin). Although it is possible that these compounds work synergistically in their function as alternative herbs. However, perhaps the mechanism of action of these compounds can be evaluated through predictions of interactions between compounds in silico. As illustrated in Fig. S1,† these compounds interact with many proteins associated with cell proliferation, including EGFR and CASP3. Numerous studies indicate that EGFR activation may be suppressed by quercetin,41 rutin,42 and quercetin 3-beta-D-glucoside43 which these compounds have been shown to activate Caspase.44Furthermore, we introduced bioactive components of Meniran into ZIF-8 nanoparticles at ambient temperature and evaluated them using a spectrophotometer with absorbance at 285 nm to precisely quantify of encapsulation efficiency. Meniran contains bioactive compounds such as flavonoids, tannins, and phenolic groups, which are capable of forming weak coordination interactions with Zn2+ in an aqueous system.45,46 As illustrated in Table S2†, the Meniran-incorporated ZIF-8 at a weight ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 attained the maximum drug-loading content (DLC) of 5.01 ± 0.85%. These results consistent with previous studies that MOF including ZIF-8 has high payload capacity.47–49
1 attained the maximum drug-loading content (DLC) of 5.01 ± 0.85%. These results consistent with previous studies that MOF including ZIF-8 has high payload capacity.47–49
The structure of ZIF-8, Meniran and Meniran-incorporated ZIF-8 was initially examined by Fourier Transform Infra-Red (FTIR) and X-ray diffraction (XRD). As shown in Fig. 1a, the distinctive peaks from IR spectroscopy of ZIF-8 are located in the 900–1350 cm−1 (bending of the imidazole ring inside the plane), <800 cm−1 (bending of the imidazole ring out of the plane) and peak around 420 cm−1, implying the existence of a chemical bond between the metal and the ligand. The primary peaks of Meniran extract are seen at 3305 cm−1 (O–H stretching), 2942 cm−1 (CH2– stretching), 2854 cm−1 (–CH3 stretching), 1611 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds), 1440 cm−1 and 1332 cm−1 (–CH3 twisting and wagging), and 1021 cm−1 (C–O stretching in secondary alcohols). The Meniran-incorporated ZIF-8 has similar peaks to pristine ZIF-8, indicating that are several overlapping peak locations between Meniran and ZIF-8. Moreover, the X-ray powder diffraction (XRD) patterns verified that samples were crystalline ZIF-8 in the I43m space group, exhibiting distinct and intense diffraction peaks that align with previous studies 50–52 (Fig. 1b). The diffractogram of Meniran indicates that partly amorphous structure with highest peak at 2θ = 69.6°, while Meniran-incorporated ZIF-8 did not exhibit any altered peak from the diffractogram of pure ZIF-8 thereby enabling the MOF to preserve its intrinsic structural characteristics. Notably, a small peak was noticed at about 2θ = 69.6° (shown by a grey arrow), which suggests the existence of Meniran within ZIF-8.
C bonds), 1440 cm−1 and 1332 cm−1 (–CH3 twisting and wagging), and 1021 cm−1 (C–O stretching in secondary alcohols). The Meniran-incorporated ZIF-8 has similar peaks to pristine ZIF-8, indicating that are several overlapping peak locations between Meniran and ZIF-8. Moreover, the X-ray powder diffraction (XRD) patterns verified that samples were crystalline ZIF-8 in the I43m space group, exhibiting distinct and intense diffraction peaks that align with previous studies 50–52 (Fig. 1b). The diffractogram of Meniran indicates that partly amorphous structure with highest peak at 2θ = 69.6°, while Meniran-incorporated ZIF-8 did not exhibit any altered peak from the diffractogram of pure ZIF-8 thereby enabling the MOF to preserve its intrinsic structural characteristics. Notably, a small peak was noticed at about 2θ = 69.6° (shown by a grey arrow), which suggests the existence of Meniran within ZIF-8.
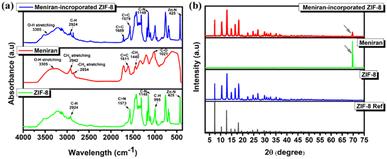 | ||
| Fig. 1 Characteristic of ZIF-8, Meniran, and Meniran-incorporated ZIF-8 under (a) FTIR spectra and (b) XRD pattern. | ||
To examine the surface structure of the samples, a field emission scanning electron microscopy (FE-SEM) was performed equipping with an energy dispersive spectroscopy at a working voltage of 20 kV. As shown in Fig. 2a, the morphology of ZIF-8 is well-distribution as nanostructure with rhombic dodecahedrons, polyhedrons with 12 faces, 24 edges and 14 vertices as agreement with previous studies. 49 Interestingly, Meniran-incorporated ZIF-8 has almost unchanged the morphology, indicating that ZIF-8 maintained the morphology of nanostructure (Fig. 2b). Furthermore, TEM analysis confirmed that the Meniran-incorporated ZIF-8 exhibited rhombic dodecahedron shapes and were slightly larger than the pristine ZIF-8 nanoparticles †. The particle size is a critical element in the effectiveness of tumor cell therapies, ZIF-8 nanoparticles with a size of 70 nm can successfully internalize towards tumor cells.53 To verify these results, we further evaluated the particle size distribution by DLS. As shown in Fig. 2c, the particle size of ZIF-8 nanoparticles is 76.2 ± 1.98, while Meniran-incorporated ZIF-8 nanoparticles exhibited particle size slightly higher of 85.1 ± 1.05. These findings indicate that the increasing particle size might be due to Meniran already payload into ZIF-8 nanoparticles. Next, to clearly understand of surface area of the ZIF-8 nanoparticles and Meniran-incorporated ZIF-8 nanoparticles, we examined by Brunauer–Emmett–Teller BET analysis of nitrogen adsorption isotherms. As shown in Fig. 2d, the surface area of ZIF-8 nanoparticles was determined to be 1021 m2 g−1 and pore volume 0.43 cm3 g−1, which is consistent with other reports with a range from 960 to 1820 m2 g−1. 54 However, upon the incorporation of Meniran into ZIF-8 nanoparticles, the surface area significantly decreased up to 403 m2 g−1 with pore volume 0.11 cm3 g−1, indicating that may be attributed to Meniran occupying the adsorption sites of the ZIF-8 nanoparticles while the smaller change may be attributed to the specific adsorption mechanism involved. These results are in excellent agreement with SEM and TEM result.
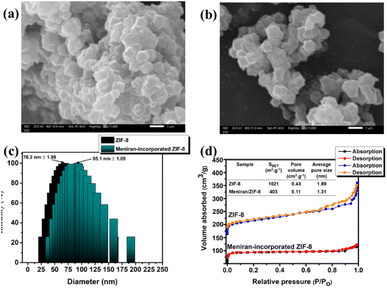 | ||
| Fig. 2 SEM images of (a) ZIF-8 and (b) Meniran-incorporated ZIF-8, (c) DLS spectra, and (d) BET analysis of ZIF-8 and Meniran-incorporated ZIF-8. | ||
Based on previous reports ZIF-8 has pH-responsive properties that have inspired us to investigate the in vitro drug release kinetics under PBS conditions. As depicted in Fig. 3a, under pH 7.4 of Meniran-incorporated ZIF-8 nanoparticles only 28% released after 36 h. Importantly, under pH 5.0 Meniran was rapidly released reached 70% and restrained release after 36 h. The Meniran-incorporated ZIF-8 exhibits pH-responsive release capacity due to the coordination bond disruption between zinc ions and imidazolate at lower pH levels. 55 Thus, the acidic environment surrounding malignancy can provide the stimulation to trigger the rapid release of Meniran from the carriers.
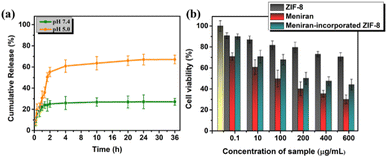 | ||
| Fig. 3 (a) In vitro drug release of Meniran-incorporated ZIF-8 nanoparticles under different pH values. (b) MTT analysis of ZIF-8, Meniran, and Meniran-incorporated ZIF-8 under HeLa cells. | ||
Furthermore, we performed a cytotoxicity assay of ZIF-8, Meniran, and Meniran-incorporated ZIF-8 nanoparticles toward HeLa cells due a stable and reproducible results across multiple experiments by MTT method. As shown in Fig. 3b, the cells viability only slightly decreases following incubation with ZIF-8 nanoparticles and without any IC50 value, suggesting that these nanocarriers are biocompatible. Surprisingly, Meniran-incorporated ZIF-8 exhibited rapidly released and cell viability significantly decreased with IC50 = 205.41 ± 3.09 μg mL−1), compared with pure Meniran IC50 102.37 ± 2.89 μg mL−1 indicating that the time required for Meniran to be released from ZIF-8 nanoparticles and consequently delayed of Meniran internalization towards the cells. Therefore, these findings confirm that bioactive compound from Meniran extract incorporated into ZIF-8 nanoparticles potent cytotoxicity in cancer cells and improve efficacy chemotherapy.
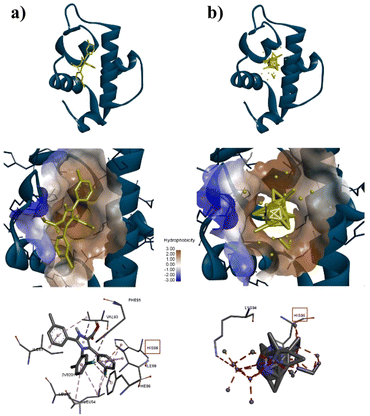
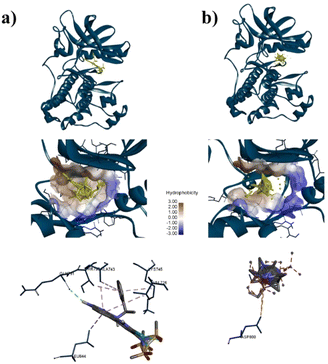
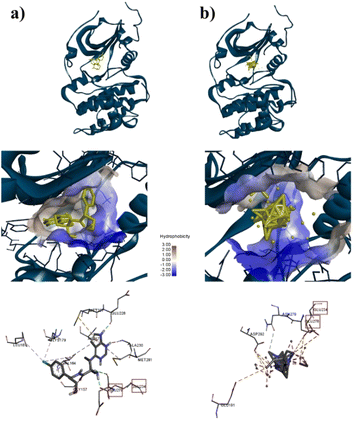
To confirm the adsorption of ZIF-8 could has interact with protein target for anticancer, molecular docking calculations were implemented. As shown in Table S3†, the docking results for AKT-1, EGFR, or MDM2 with ZIF-8 demonstrated higher binding affinities to AKT-1 and EGFR compared to a known inhibitor, with affinities of −10.8 kcal mol−1 versus AZD5363's −9.1 kcal mol−1 for AKT-1, and −8.5 kcal mol−1 versus MTX-531's −8.2 kcal mol−1 for AKT-1. However, in MDM2, ZIF-8 is potentially bound with binding affinities of −9.4 kcal mol−1, compared inhibitor (Imidazole, −9.6 kcal mol−1). Furthermore, we conducted an investigation focused on assessing the alignment of the compounds with the protein active site owing to crucial for evaluating the potential of these nanoparticle to inhibit the proteins effectively. The molecular docking analysis revealed that ZIF-8 nanoparticles could bind to a similar region as the inhibitor (Fig. 4–6). However, only in the cases of MDM2 and AKT-1 did ZIF-8 exhibit comparable amino acid residue interactions to those of the control ligand (Fig. 4 and 5). The ZIF-8 nanoparticles binding to HIS96 is comparable to that of the native inhibitor when interacting with the MDM2 protein (Fig. 4). Similarly, ZIF-8 binding to GLU234 and GLU278 mirrors the native inhibitor's interactions with the MDM2 protein (Fig. 5).
Nanoparticles (NPs) often exhibit distinct magnetic, thermal, optical, and electrical properties due to their large surface area and significant quantum mechanical effects. They are frequently designed and employed as drug carriers, facilitating the targeted delivery of chemotherapeutic agents to tumor tissues while reducing damage to healthy organs. 56 ZIF-8 is a type of nanoparticle, composed of zinc metal and 2-methyl imidazole (MIM) ligand, are one of the most widely used types of ZIFs owing to their inherent properties, such as the low toxicity of zinc metal, relatively large cavities, high thermal and chemical stability, ultrahigh surface area, and high crystallinity. 57 Moreover, the LD50 (the dosage at which 50% of the animal population is killed) for ZIF-8 has been documented as 1400 mg mL−1. 57,58 This research aimed to assess the effectiveness of ZIF-8 as a drug nanocarrier for anticancer agents by molecular docking analysis.
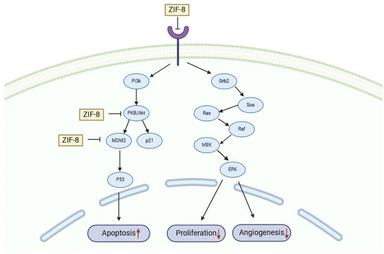
Next, our study used three specific proteins, namely AKT-1, MDM2, and EGFR which are implicated in cancer pathways, such as regulating cell proliferation, angiogenesis, and apoptosis. Several studies have concentrated on targeting these proteins in cancer therapy. 59–63 Based on Table S3†, ZIF-8 nanoparticles have potential to binding with three proteins above which binding affinity as same as the native inhibitor. Then, ZIF-8 nanoparticles have comparable interactions with amino acid residues when compared to the control ligand. These interactions play a role in the anticancer pathway by inhibiting AKT-1, MDM2, and EGFR. Suppression of both proteins may result in reduced cell growth and formation of new blood vessels, which can ultimately lead to cell death (Fig. 7). Hence, further in vitro study is required to validate the efficacy of the drugs being studied.
Conclusion
The bioactive compound was extracted from the leaves of Meniran (Phyllanthus niruri L.) using methanol and successfully identified through LC-HRMS analysis. Subsequently, Meniran was incorporated into ZIF-8 to form nanoparticles, and FTIR and XRD analyses were conducted to confirm the chemical structure. The resulting Meniran-incorporated ZIF-8 nanoparticles successfully maintained their chemical and crystalline structure, compared to pure ZIF-8. Moreover, the shape of Meniran-incorporated ZIF-8 nanoparticles shows well-distribution nanoparticles with rhombic dodecahedrons which provide to internalize towards cancer cells. The Meniran-incorporated ZIF-8 nanoparticles showed rapidly release under acidic environment while slowly release under normal condition. Interestingly, in vitro analysis showed that Meniran-incorporated ZIF-8 nanoparticles has anticancer activity towards HeLa cells. To support these results, computational simulations clearly demonstrated that ZIF-8 as nanocarrier has molecular-level interaction with EGFR, AKT-1, MDM-2 under cancer cells. Thus, this study provides new insight to development of bioactive compounds encapsulated into nanoparticles as cancer treatment.Experimental section
Materials
Phyllanthus niruri (Meniran) was sourced from a local plant in East Java, Indonesia. Zinc nitrate hexahydrate (Zn(NO3)2·6H2O, 98%), 2-methylimidazole (C4H6N2, 99%) were purchased from Sigma-Aldrich (USA). Ultrapure water, purified using Milli-Q Plus 185 equipment, was consistently utilized in all experimental procedures throughout this study.Meniran extraction process
Phyllanthus niruri L. (Meniran) leaves were obtained from Mount Lawu at an altitude of 914 masl with coordinates 7°34′00′′S 111°13′16′′E. A total of ±1 kg of dry leaf powder of Meniran was extracted by maceration method using methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 b/v). Extraction was carried out for 1 × 24 hours and repeated 3 times. The extract of Meniran was concentrated with a rotary vacuum evaporator at 45 °C (Buchi Rotavapor R-100).
3 b/v). Extraction was carried out for 1 × 24 hours and repeated 3 times. The extract of Meniran was concentrated with a rotary vacuum evaporator at 45 °C (Buchi Rotavapor R-100).
Synthesis ZIF-8 nanoparticles
ZIF-8 nanoparticles were synthesized following the methodology outlined in a previous report. 36,37 2-Methylimidazole (5.67 g) was dissolved in 20 mL of water, while zinc nitrate hexahydrate (0.29 g) was dissolved in 2 mL of water. Subsequently, the aqueous zinc nitrate solution (0.29 g, 2 mL) was added to the aqueous 2-methylimidazole solution and stirred vigorously for 4 h at room temperature. Finally, the milky white precipitates were washed several times with water through centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm. The resulting product was then dried in an oven at 40 °C for 24 h.
500 rpm. The resulting product was then dried in an oven at 40 °C for 24 h.
Preparation incorporation of Meniran into ZIF-8 nanoparticle
A mixture of 30 mg of Meniran extract and 10 mg of ZIF-8 in 5 mL of methanol was stirred for 48 h at room temperature, with the vial kept sealed to prevent methanol evaporation. The powder was dried at room temperature for 24 h. The supernatant obtained after centrifugation was used to quantify the amount of Meniran loaded.Active compound analysis using liquid chromatography high resolution mass spectrometry (LC-HRMS)
LC-HRMS analysis was performed at the Central Laboratory of Life Sciences, Brawijaya University. Sample preparation was carried out by dissolving the combination of extracts in water solvent. The clear extract solution was injected into LC-HRMS (Thermo Scientific Dionex Ultimate 3000 RSLCnano with microflow meter). The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Chromatographic separation was performed with a Hypersil GOLD aQ analytical column particle size of 50 × 1 mm × 1.9 μm with an analytical flow rate of 40 μL min−1. The column temperature was set at 40 °C, and the running time was 30 minutes. The polarity was adjusted to either positive or negative. Full scan at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and data-dependent MS2 was at a resolution of 17
000, and data-dependent MS2 was at a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. Data processing using Compound Discoverer (Thermo Scientific) with McCloud MS/MS Library.
500. Data processing using Compound Discoverer (Thermo Scientific) with McCloud MS/MS Library.
Release of Meniran from Meniran-incorporated ZIF-8 nanoparticles under different pH
A standard release system was prepared in PBS solution at varying pH levels of 7.4 and 5.0. The release system was subsequently maintained at 25 °C with continuous stirring, and 3 mL of the release medium was withdrawn at each designated time point. The supernatant was collected through centrifugation and utilized to quantify the concentration of released Meniran using UV-vis spectroscopy with absorbance at 285 nm.Characterization
Cytotoxicity test
HeLa cells were seeded into 96-well plates at a density of 1 × 105 cells per well in 100 μL of medium. The medium was substituted with a fresh one containing pristine ZIF-8 and Meniran-incorporated ZIF-8 at various concentrations, subsequently culture for 24 h. Then, 20 μL of MTT reagent was introduced into each well and incubated for 4 h. Finally, 100 μL of DMSO was added to each well to dissolve the formazan generated by MTT and measured using an ELISA microplate reader.Computational simulations
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.† Raw data that support the findings of this study are available from the corresponding author, upon reasonable request.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was supported financially by Ministry of Research, Technology, and Higher Education Republic of Indonesia (DRTPM) (contract no. B/61648/UN38.III.1/LK.04.00/2024).References
- Y. Octavia, C. G. Tocchetti, K. L. Gabrielson, S. Janssens, H. J. Crijns and A. L. Moens, J. Mol. Cell. Cardiol., 2012, 52, 1213–1225 CrossRef CAS.
- Y. Cui, J. Sui, M. He, Z. Xu, Y. Sun, J. Liang, Y. Fan and X. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 2193–2203 CrossRef CAS PubMed.
- Y. B. Lim, K. S. Moon and M. Lee, Chem. Soc. Rev., 2009, 38, 925–934 RSC.
- J. S. Lee and J. Feijen, J. Controlled Release, 2012, 161, 473–483 CrossRef CAS.
- Z. Zhang, X. Chen, L. Chen, S. Yu, Y. Cao, C. He and X. Chen, ACS Appl. Mater. Interfaces, 2013, 5, 10760–10766 CrossRef CAS PubMed.
- X. Guo, D. Li, G. Yang, C. Shi, Z. Tang, J. Wang and S. Zhou, ACS Appl. Mater. Interfaces, 2014, 6, 8549–8559 CrossRef CAS PubMed.
- H. Li, Y. Cui, J. Sui, S. Bian, Y. Sun, J. Liang, Y. Fan and X. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 15855–15865 CrossRef CAS.
- J. Fang, H. Nakamura and H. Maeda, Adv. Drug Delivery Rev., 2011, 63, 136–151 CrossRef CAS.
- D. Jiang, W. Mu, X. Pang, Y. Liu, N. Zhang, Y. Song and S. Garg, ACS Appl. Mater. Interfaces, 2018, 10, 37797–37811 CrossRef CAS PubMed.
- S. Ganta, H. Devalapally, A. Shahiwala and M. Amiji, J. Controlled Release, 2008, 126, 187–204 CrossRef CAS PubMed.
- D. A. T. ATEŞ, ÖZLEM Turkish Journal of Biology, 2003, 27, 157–162 Search PubMed.
- W. Nurcholis, B. P. Purwakusumah, E. D. Purwakusumah, T. Katayama and T. Suzuki, J. Kim. Valensi, 2012, 2, 501–510 Search PubMed.
- S.-T. Huang, R.-C. Yang, L.-J. Yang, P.-N. Lee and J.-H. S. Pang, Life Sci., 2003, 72, 1705–1716 CrossRef CAS PubMed.
- H.-H. Tseng, P.-N. Chen, W.-H. Kuo, J.-W. Wang, S.-C. Chu and Y.-S. Hsieh, Integr. Cancer Ther., 2011, 11, 267–278 CrossRef PubMed.
- S.-T. Huang, R.-C. Yang, M.-Y. Chen and J.-H. S. Pang, Life Sci., 2004, 75, 339–351 CrossRef CAS.
- N. Chudapongse, M. Kamkhunthod and K. Poompachee, J. Ethnopharmacol., 2010, 130, 315–319 CrossRef.
- H.-Y. Wu, T.-K. Lin, H.-M. Kuo, Y.-L. Huang, C.-W. Liou, P.-W. Wang, J.-H. Chuang and S.-T. Huang, J. Evidence-Based Complementary Altern. Med., 2012, 2012, 925824 Search PubMed.
- S. H. Lee, I. B. Jaganath, R. Manikam and S. D. Sekaran, BMC Compl. Alternative Med., 2013, 13, 271 CrossRef PubMed.
- M. Geethangili and S. T. Ding, Front. Pharmacol, 2018, 9, 1109 CrossRef CAS PubMed.
- J. Voigt, Food/Nahrung, 1993, 37, 185 Search PubMed.
- W. Liang, P. Wied, F. Carraro, C. J. Sumby, B. Nidetzky, C.-K. Tsung, P. Falcaro and C. J. Doonan, Chem. Rev., 2021, 121, 1077–1129 CrossRef CAS.
- L. Jiao, J. Y. R. Seow, W. S. Skinner, Z. U. Wang and H.-L. Jiang, Mater. Today, 2019, 27, 43–68 CrossRef CAS.
- H. D. Lawson, S. P. Walton and C. Chan, ACS Appl. Mater. Interfaces, 2021, 13, 7004–7020 CrossRef CAS.
- N. V. de Almeida Ferraz, W. Silva Vasconcelos, C. Santos Silva, S. Alves Junior, C. G. Amorim, M. da Conceição, B. S. M. Montenegro and M. C. da Cunha Areias, Sens. Actuators, B, 2020, 307, 127636 CrossRef.
- J. Rosário, L. L. da Luz, R. Geris, J. G. S. Ramalho, A. F. da Silva, S. A. Júnior and M. Malta, Sci. Rep., 2019, 9, 7302 CrossRef PubMed.
- S. G. F. de Assis, G. C. Santos, A. B. S. Santos, E. H. L. Falcão, R. da Silva Viana and S. A. Junior, J. Solid State Chem., 2019, 276, 309–318 CrossRef CAS.
- E. Aghazadeh Asl, M. Pooresmaeil and H. Namazi, Mater. Chem. Phys., 2023, 293, 126933 CrossRef CAS.
- Q. Wang, Y. Sun, S. Li, P. Zhang and Q. Yao, RSC Adv., 2020, 10, 37600–37620 RSC.
- X. Dou, M. Keywanlu, R. Tayebee and B. Mahdavi, J. Mol. Liq., 2021, 329, 115557 CrossRef CAS.
- I. B. Vasconcelos, T. G. d. Silva, G. C. G. Militão, T. A. Soares, N. M. Rodrigues, M. O. Rodrigues, N. B. d. Costa, R. O. Freire and S. A. Junior, RSC Adv., 2012, 2, 9437–9442 RSC.
- J. S. F. Silva, J. Y. R. Silva, G. F. de Sá, S. S. Araújo, M. A. G. Filho, C. M. Ronconi, T. C. Santos and S. A. Júnior, ACS Omega, 2018, 3, 12147–12157 CrossRef CAS.
- L. R. de Moura Ferraz, A. É. G. A. Tabosa, D. D. S. da Silva Nascimento, A. S. Ferreira, V. de Albuquerque Wanderley Sales, J. Y. R. Silva, S. A. Júnior, L. A. Rolim, J. J. de Souza Pereira and P. J. Rolim-Neto, Sci. Rep., 2020, 10, 16815 CrossRef CAS PubMed.
- S. Feng, X. Zhang, D. Shi and Z. Wang, Front. Chem. Sci. Eng., 2021, 15, 221–237 CrossRef CAS.
- C.-Y. Sun, C. Qin, X.-L. Wang, G.-S. Yang, K.-Z. Shao, Y.-Q. Lan, Z.-M. Su, P. Huang, C.-G. Wang and E.-B. Wang, Dalton Trans., 2012, 41, 6906–6909 RSC.
- H. Ren, L. Zhang, J. An, T. Wang, L. Li, X. Si, L. He, X. Wu, C. Wang and Z. Su, Chem. Commun., 2014, 50, 1000–1002 RSC.
- J. Redfern, L. Geerts, J. W. Seo, J. Verran, L. Tosheva and L. H. Wee, ACS Appl. Nano Mater., 2018, 1, 1657–1665 CrossRef CAS.
- T. Imae, A. Rahmawati, A. M. Berhe and M. A. Kebede, ACS Appl. Nano Mater., 2022, 5, 16842–16852 CrossRef CAS.
- O. Aung Myo, N. Mohd Nasir Mat, L. Ohn Mar and S. Nordin, Asian Journal of Medicine and Biomedicine, 2022, 6, 17–31 Search PubMed.
- D. Tungmunnithum, A. Thongboonyou, A. Pholboon and A. Yangsabai, Medicine, 2018, 5, 93 CAS.
- N. Q. Nguyen, L. V. Minh, L. H. Trieu, L. M. Bui, T. D. Lam, V. Q. Hieu, T. V. Khang and L. N. Y. Trung, IOP Conf. Ser.:Mater. Sci. Eng., 2020, 736, 062017 CAS.
- A. B. Firdous, G. Sharmila, S. Balakrishnan, P. RajaSingh, S. Suganya, N. Srinivasan and J. Arunakaran, Food Funct., 2014, 5, 2632–2645 RSC.
- J. Lee, J. Lee, W. Sim, J. H. Kim, C. Choi and J. Jeon, BMB Rep., 2023, 56, 594–599 CrossRef CAS.
- N. Lotfi, Z. Yousefi, M. Golabi, P. Khalilian, B. Ghezelbash, M. Montazeri, M. H. Shams, P. Z. Baghbadorani and N. Eskandari, Front. Immunol., 2023, 14, 1077531 CrossRef CAS PubMed.
- Q. Chen, P. Li, P. Li, Y. Xu, Y. Li and B. Tang, Oncol. Rep., 2023, 49, 6 Search PubMed.
- L. Xing, H. Zheng, Y. Cao and S. Che, Adv. Mater., 2012, 24, 6433–6437 CrossRef CAS.
- F. Novio, J. Simmchen, N. Vázquez-Mera, L. Amorín-Ferré and D. Ruiz-Molina, Coord. Chem. Rev., 2013, 257, 2839–2847 CrossRef CAS.
- K. Jiang, L. Zhang, Q. Hu, X. Zhang, J. Zhang, Y. Cui, Y. Yang, B. Li and G. Qian, Microporous Mesoporous Mater., 2019, 275, 229–234 CrossRef CAS.
- G. Chen, J. Luo, M. Cai, L. Qin, Y. Wang, L. Gao, P. Huang, Y. Yu, Y. Ding, X. Dong, X. Yin and J. Ni, Molecules, 2019, 24, 3369 CrossRef CAS.
- W. Cai, J. Wang, C. Chu, W. Chen, C. Wu and G. Liu, Advanced Science, 2019, 6, 1801526 CrossRef PubMed.
- K. Qiu, Y. Shu, J. Zhang, L. Gao and G. Xiao, Catal. Lett., 2022, 152, 172–186 CrossRef CAS.
- D. W. Lewis, A. R. Ruiz-Salvador, A. Gómez, L. M. Rodriguez-Albelo, F.-X. Coudert, B. Slater, A. K. Cheetham and C. Mellot-Draznieks, CrystEngComm, 2009, 11, 2272–2276 RSC.
- M. He, J. Yao, Q. Liu, K. Wang, F. Chen and H. Wang, Microporous Mesoporous Mater., 2014, 184, 55–60 CrossRef CAS.
- X. Liu, Y. Li, Y. Ban, Y. Peng, H. Jin, H. Bux, L. Xu, J. Caro and W. Yang, Chem. Commun., 2013, 49, 9140–9142 RSC.
- J. Zhuang, C.-H. Kuo, L.-Y. Chou, D.-Y. Liu, E. Weerapana and C.-K. Tsung, ACS Nano, 2014, 8, 2812–2819 CrossRef CAS.
- D. N. Ta, H. K. D. Nguyen, B. X. Trinh, Q. T. N. Le, H. N. Ta and H. T. Nguyen, Can. J. Chem. Eng., 2018, 96, 1518–1531 CrossRef CAS.
- F.-S. Liao, W.-S. Lo, Y.-S. Hsu, C.-C. Wu, S.-C. Wang, F.-K. Shieh, J. V. Morabito, L.-Y. Chou, K. C. W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2017, 139, 6530–6533 CrossRef CAS PubMed.
- X. Yu, I. Trase, M. Ren, K. Duval, X. Guo and Z. Chen, J. Nanomater., 2016, 2016, 1087250 Search PubMed.
- M. Ahmadi, M. Khoramjouy, S. Dadashzadeh, E. Asadian, M. Mosayebnia, P. Geramifar, S. Shahhosseini and F. Ghorbani-Bidkorpeh, J. Drug Delivery Sci. Technol., 2023, 81, 104249 CrossRef CAS.
- M. A. Elaziz, N. S. Abdelrahman, N. A. Hassan and M. O. Mohamed, Math. Probl. Eng., 2022, 2022, 3029932 Search PubMed.
- L. Chibaya, B. Karim, H. Zhang and S. N. Jones, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2003193118 CrossRef CAS.
- Z. Tóthová, M. Šemeláková, Z. Solárová, J. Tomc, N. Debeljak and P. Solár, Int. J. Mol. Sci., 2021, 22, 7682 CrossRef.
- B. George, B. Gui, R. Raguraman, A. M. Paul, H. Nakshatri, M. R. Pillai and R. Kumar, Cells, 2022, 11, 2290 CrossRef CAS PubMed.
- T. L. Wargasetia, H. Ratnawati, N. Widodo and M. H. Widyananda, Cancer Inf., 2021, 20, 11769351211031864 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: LC-HRMS analysis, loading content, binding affinity results are provided. See DOI: https://doi.org/10.1039/d4ra06399f |
| This journal is © The Royal Society of Chemistry 2025 |