Open Access Article
Open Access ArticleMetal compounds as antimicrobial agents: ‘smart’ approaches for discovering new effective treatments
Valentina Vitali†
 ,
Stefano Zineddu†
,
Stefano Zineddu† and
Luigi Messori
and
Luigi Messori *
*
Department of Chemistry “Ugo Schiff”, University of Florence, Sesto Fiorentino, 50019, Italy. E-mail: luigi.messori@unifi.it
First published on 9th January 2025
Abstract
Due to their considerable chemical diversity, metal compounds are attracting increasing and renewed attention from the scientific and medical communities as potential antimicrobial agents to combat the growing problem of antibiotic resistance. The development of metal compounds as antimicrobial agents typically follows classical drug discovery procedures and suffers from the same problems; indeed, these procedures can be very expensive and time-consuming, and carry an intrinsically high risk of failure. Here, we show how some established drug discovery approaches can be conveniently and successfully applied to antimicrobial metal compounds to provide some shortcuts for faster clinical translation of new treatments. Specifically, we refer to (i) drug repurposing, (ii) drug combination and (iii) drug targeting by bioconjugation; some relevant examples will be illustrated.
1. Introduction
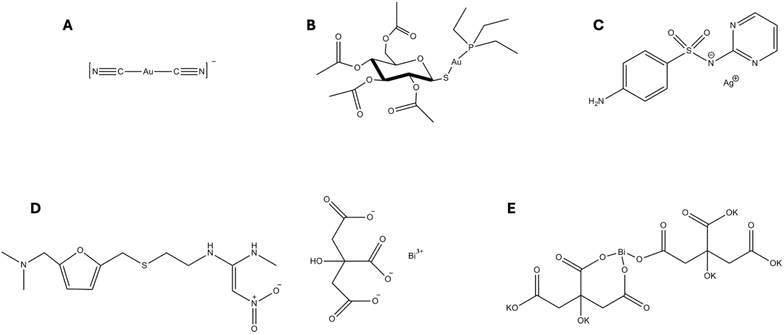
Metals are essential for the survival of all living organisms, but different forms or exposure to higher concentrations of the same metal can cause severe toxicity. In analogy to eukaryotic systems, some bacterial enzymes contain specific metals that catalyse essential biochemical reactions. To maintain proper function, bacterial cells meticulously regulate the homeostasis of these metals: excesses are avoided to prevent toxicity, while at the same time insufficient levels can impair cellular requirements.1For centuries, a wide range of metals and metal-based compounds have been used empirically as anti-infective agents with remarkable results.2 In particular, the gold complex dicyanoaurate(I) (Fig. 1A) was proposed by Robert Koch as an antitubercular agent in the pioneering days of modern pharmacology.3 Similarly, several bismuth, antimony and mercury compounds have been used therapeutically to treat various bacterial and parasitic diseases.4–6 However, the advent of the golden age of antibiotics, coupled with legitimate concerns about their systemic toxicity, led to the gradual abandonment of metal-based agents and their restricted clinical use.
Despite their historic importance in modern medicine, the efficacy of antibiotics has been increasingly threatened in recent decades by the relentless development of resistance among bacterial pathogens. Such resistance is mainly imputable to the misuse and abuse of antibiotics in agriculture and intensive farming practices,7 as well as to the excessive and often unnecessary prescriptions to treat common flu symptoms. In addition, inappropriate choice of dose and duration of therapy often only leads to partial elimination of pathogens, inadvertently promoting the development of resistance. These resistance mechanisms can offer bacteria the chance to develop evasion mechanisms against multiple classes of antibiotics, resulting in multi-drug resistant (MDR) or pan-drug resistant (PDR) phenotypes that mirror situations typical of the pre-antibiotic era. These phenomena have triggered the so-called “antimicrobial resistance crisis”, a critical global challenge that requires immediate attention.
It is predicted that infectious diseases will cause more deaths than cancer worldwide within the next 25 years,8 while the number of newly approved and marketed antibiotics has significantly declined since the late 1970s. There is therefore a dramatic need for the discovery of novel antimicrobials and therapeutic strategies to combat life-threatening infections caused by resistant pathogens. In this context, the vast chemical diversity of metal-based compounds has attracted renewed interest towards them as a promising source of novel synthetic antimicrobial agents with unique and unprecedented mechanisms of action. Although only bismuth and silver-based antimicrobials (Fig. 1C–E) are currently in clinical use, the immense versatility of metal-based compounds, including the types of ligands, the coordination geometries and the intrinsic properties of the metallic centre, make them some of the most promising antibacterial candidates for the next generation of antimicrobials.
2. Metal-based compounds as antimicrobials
Regardless of their long history as antibacterial agents, only two families of metal-derived compounds, bismuth and silver, are currently used in the clinical treatment of bacterial infections. Bismuth has broad-spectrum activity and can be administered in various formulations, either alone or in combination with other antibiotics, to treat a wide range of conditions including Helicobacter pylori infections,9 wound infections,10 and diabetic foot ulcers.11 Similarly, silver compounds find widespread application in wound healing and management of infections.9 However, the potential of metal-based compounds as a source of novel antimicrobial agents remains largely underestimated and underexplored, despite the immense versatility offered by inorganic coordination chemistry, which could potentially overcome the limitations of purely organic molecules. Only recently has there been a growing interest in these peculiar compounds. Several reviews on the state of the art of metal compounds as antimicrobials have been published.11,12 One of these studies evaluated and compared the antimicrobial activity of a vast library (nearly one thousand) of metal complexes against the ESKAPE (acronym for E. faecium, S. aureus, K. pneumoniae, A. baumanii, P. aeruginosa and Enterobacter species) panel of pathogens, detecting some peculiar trends.11 Indeed, among the screened metal compounds with acceptable toxicity profiles, 9.9% showed promising activity, compared to a mere 0.87% for purely organic compounds (considering as active the compounds with Minimal Inhibitory Concentration values equal or lower to 16 μg mL−1 against at least one pathogen of the panel). This surprisingly high success rate of more than 10-fold is probably due to the variety of geometries and ligand types that can be found around the central metal atom. Furthermore, unlike organic molecules, which are primarily confined to one- and two-dimensional chemical space, metal compounds offer access to unexplored three-dimensional chemical space, making these compounds highly attractive for the future development of antimicrobial agents.13In addition, metal complexes have unique mechanisms of action that are difficult to achieve with purely organic substances. These compounds typically exert their antibacterial activity through two different modes: either the entire metal complex is required for the antimicrobial activity, or the activity is solely mediated by the metal ion. In the first mode, the mechanism of action depends critically on the specific metal within the complex, including its oxidation state, its geometrical arrangement and the nature of the ligands. For example, Au(I) and Au(III) compounds have been shown to be effective inhibitors of bacterial thioredoxin reductase (TrxR), but have also been implicated in off-target activity on bacterial cell wall and protein synthesis. In contrast, some Ru(II) complexes have demonstrated antibacterial activity through membrane disruption and inhibition of biofilms formation,14,15 while piano-stool Ru(II) complexes are believed to act as intercalating DNA binding agents.16 Overall, the versatility of metal-based complexes allows for a wide range of mechanisms of action, often involving the generation of Reactive Oxygen Species (ROS), ligand release or exchange reactions, and activation of electron transfer processes (redox activation).17,18 On the other hand, when acting in the second mode, metal ions can dissociate from the ligands and replace essential metal cofactors in the bacterial metalloenzymes. This substitution may disrupt the enzyme's catalytic activity, inhibiting crucial metabolic pathways and ultimately leading to bacterial cell death. Several examples of this behaviour have been reported during the past years involving different types of metal ions, including Au(I), Ag(I), Ga(III) and Bi(III).19 In general, the specific mechanism employed by a given metal complex depends on its unique composition, including the metal ion, the ligands and their interactions. This diversity offers potential advantages in the fight against antimicrobial resistance, as bacteria may find it difficult to develop resistance to multiple mechanisms of action. Below, we will consider some effective “smart” strategies for the development and use of metal compounds in the treatment of infectious diseases; specifically, we will refer to three different approaches: (i) drug repurposing, (ii) combination therapies and (iii) drug targeting through bioconjugation, all applied to antimicrobial metal compounds.
3. Drug repurposing
Traditional drug development and approval processes involve multiple steps ranging from synthesis to market launch, through multiple clinical trials; such processes are often time-consuming, very expensive and riddled with failures and missteps. The success rate at the end of the process is extremely low.Recently, the concept of drug repurposing has emerged as a strategy to overcome the significant financial risks associated with traditional drug development. Drug repurposing involves testing existing, FDA-approved drugs for new uses beyond their original therapeutic indications. This approach offers significant advantages because the safety profiles and pharmacological properties of these drugs are already well established.
Gallium nitrate (Ganite, Ga(NO3)3) represents a notable example of metal-based drug repurposing. Initially approved for the treatment of hypercalcemia in cancer patients, this compound has recently demonstrated significant antibacterial and antibiofilm activities against the Gram-negative bacterium Pseudomonas aeruginosa, a key pathogen associated with multidrug resistance and chronic infections. It is widely accepted that Ganite exerts its antibacterial effects by mimicking the behaviour and function of iron(III) ions, disrupting bacterial iron metabolism, a critical pathway for bacterial survival and virulence, or acting as iron contender by competitively binding to iron-dependent proteins.20 Similarly, bismuth subsalicylate (BSS, C7H5BiO), a compound originally approved by the FDA for the treatment of gastrointestinal disorders such as diarrhoea, nausea, and indigestion, has recently attracted attention for its potential antimicrobial applications. This compound has demonstrated notable activity against a range of bacterial pathogens, including Clostridium difficile and Escherichia coli. The antimicrobial properties of bismuth subsalicylate are primarily attributed to its ability to disrupt bacterial cell membranes and interfere with essential enzymatic processes, particularly those involving iron and sulphur metabolism.21 However, perhaps, the most compelling example in this field is represented by the case of auranofin (AF). This Au(I) compound was approved in 1985 for the treatment of rheumatoid arthritis and nowadays is being reconsidered for a vast array of therapeutic applications, ranging from anticancer and antiviral to antifungal and antimicrobial.22–24 The latter field has gained increasing interest in recent decades, with AF undergoing extensive testing both as a single agent and in combination with other biologically relevant molecules. Promising results have been reported particularly against Gram-positive bacteria (monoderms),25 with encouraging MIC values and a positive trend even in the disruption of biofilm mass and in the fight against persister cells.26 On the other hand, AF appears less effective against Gram-negative bacteria (diderms), likely because of its difficulties in penetrating the outer membrane of the double layer.25 To address the issue of reduced drug efficacy, promising alternatives in the field of metal-based drugs are emerging as viable solutions to further improve the efficacy of antimicrobial treatments. Two main avenues are being pursued: combination therapies and drug targeting through bioconjugation. Both strategies offer distinct advantages over the use of the metallodrug alone, as discussed below.
4. Synergistic effects of combined drugs
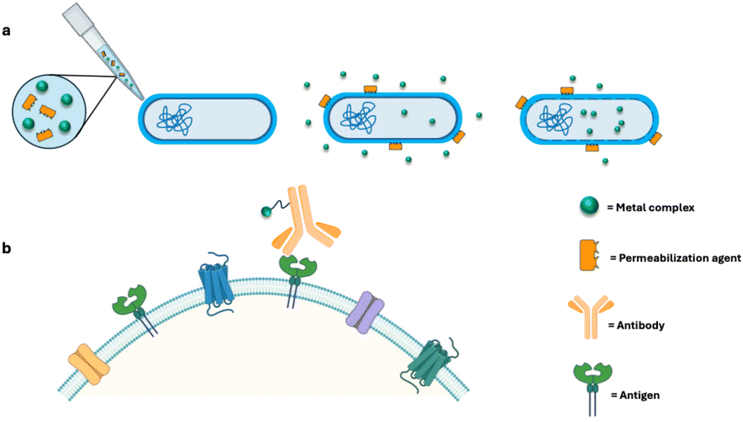
Combined therapies involve the administration of a metal-based drug with another biologically relevant molecule, such as membrane permeabilizing agents, antibiotics or other types of drugs (Fig. 2a). This strategy aims to synergistically improve the overall efficacy of the treatment. Indeed, exploiting the synergistic effects of two distinct molecules can lead to a dramatic enhancement of the antibacterial activity. This approach has already been successfully implemented in the field of antimicrobial metallodrugs by coupling the action of auranofin with that of pentamidine, an antiprotozoal drug which successfully disrupts the outer membrane of Gram-negative bacteria and increases AF intracellular uptake. The synergistic action of these two non-antibiotics significantly increases the antimicrobial performances of the gold compound, as evidenced by the reduction by one order of magnitude of the MIC values of AF.27 A similar trend was observed following the combined administration of AF and permeabilizing agent and antibiotic-derived polymyxin B nonapeptide28 (PMBN) against a wide panel of Gram-negative bacteria, again demonstrating a substantial reduction in MIC values compared to AF alone.25 Even the efficacy of traditional antibiotics can be enhanced with the presence of metal-based complexes. Linezolid and fosfomycin, combined with AF, exhibited synergistic and improved antibacterial activity when tested against Staphylococcus aureus and Enterococcus faecalis strains.29 Similar results were reported for the combination of AF with chloramphenicol improving, beyond the antimicrobial, also the anti-biofilm activity of this antibiotic compound.295. Drug targeting through bioconjugation
Drug targeting typically encompasses the administration of metallodrugs covalently linked to relevant biomolecules, mostly consisting in antibodies (Fig. 2b). This approach exploits antibody-drug conjugates (ADCs) to achieve the targeted and selective delivery of the metallodrug to specific pathogens.Through the optimization of antibody dependent therapies, it is possible to successfully increase the drug targeting selectivity of the ADC against the pathogen with respect to the free drug. Bacterial antibodies recognize specific antigens on the cell wall surface, ensuring the strategic delivery of drugs solely to bacterial pathogens. This approach offers several advantages: (i) unlike the broad spectrum antimicrobial properties of traditional antibiotics, the high selectivity of ADCs guarantees that the antibacterial action is exerted (nearly) exclusively on the pathogen, while circumventing adverse side effects on endogenous human microflora;30 (ii) at the same time, the high specificity of ADC towards specific bacterial surface proteins restricts the development of drug resistance;31 (iii) in the absence of the threat of drug resistance, lower doses of traditional antibiotics could potentially be administered.
How the conjugation between the metal complex and the antibody is carried out is the main aspect that needs to be addressed to infuse the desired features to the novel ADC system. The strategy that is typically applied employs an external linker to covalently connect the metal-based drug to a precisely selected amino acid residue on the antibody scaffold. Cysteine and lysine residues are usually the preferred targets due to the unique reactivity of the cysteine thiol group (–SH) and the extensive repertoire of chemical modifications possible with the primary amines of the lysine side chain. The selection of the conjugation site significantly influences the drug loading capacity of the ADC and subsequently its pharmacological profile. While antibodies possess a relatively abundant population of lysine residues (approximately 80 per antibody, with roughly 10 available for conjugation), cysteine residues are comparatively scarce. The most prevalent conjugation method for cysteine residues exploits a maleimide functional group which, via Michael addition with the free sulfhydryl group, forms a novel stable covalent bond. On the other hand, lysine's side chain modification usually occurs via reaction with isothiocyanates (NCS) or N-hydroxysuccinimide (NHS) esters. However, recent studies highlight the powerful role of cyclic anhydrides as a tool for this variety of biological conjugation.32
6. Challenges in the development of antimicrobial metal-based drugs
Having said all this, it is well known in the scientific community that the development of antimicrobial – and not only – metal-based drugs faces several challenges, even though the unique properties of metal centres give such compounds a significant potential. Herein, we will review some of the key issues that we, as chemists, may encounter in the design of novel metal antimicrobials.Many metal ions exhibit broad cytotoxicity against human cells. This poses a significant obstacle to the development of highly selective compounds that ensure selective targeting of microbial cells. One strategy that could be employed is the coordination to organic ligands, which can increase specificity and reduce off-target effects. A good example of this is metallophores, which are low molecular weight organic ligands that generally provide metal ion nutrients to an organism.33
Metal-based compounds often exhibit unstable behaviour in solution under physiological conditions, leading to premature degradation, poor bioavailability and complications in maintaining therapeutic levels in vivo. The use of stabilising ligands, where possible, or delivery systems could help in this situation.34,35
Another concern is microbial resistance to metal ions, which can often develop, although generally at a slower rate than for organic antibiotics. Bacterial defence systems such as efflux pumps or sequestration of metal ions may greatly reduce the efficacy of metal-based antimicrobials.36,37 To overcome this phenomenon, it is essential to design compounds that act by multiple mechanisms, making it more difficult for microbes to develop resistance. In addition, as described above, combination therapies that combine metals with other drugs (antibiotics or adjuvants) can prevent or reduce the emergence of resistance.
It is also important to consider that the mechanisms by which metal ions exert their antimicrobial effects are often complex and multifactorial, involving interactions with proteins, lipids, and nucleic acids,38 and this can make optimizing their activity challenging. Detailed mechanistic studies, leveraging techniques like proteomics, genomics, and metabolomics, can help to better understand how these metals affect microbial cells and how they might be tuned for specificity and efficacy.
7. Conclusions and future perspectives
There is an urgent need to find new drugs to combat the so-called “antibiotic resistance crisis”. Metallic compounds may offer excellent solutions in this direction, but the drug discovery process is still very long, very expensive and often fraught with failure. We have shown here how some established approaches such as drug repurposing, drug combination and drug targeting by bioconjugation can be successfully applied to antimicrobial metal compounds to accelerate the discovery and clinical translation of new effective treatments for bacterial diseases. Some successful examples have been described in detail.On the other hand, elucidating their detailed mechanism of action would be a key step in maximising the efficacy of metallic antimicrobials. The application of state-of-the-art omics technologies, such as genomics, transcriptomics, proteomics and metabolomics, could be of great help in this area. Together, these techniques can provide invaluable insights into drug–target interactions, affected biological pathways and potential downstream consequences. Therefore, the integration of omics technologies has the potential to become an essential part of antimicrobial drug discovery, both by elucidating the mechanism of action and by unravelling non-trivial response pathways that are active. Compared to traditional techniques, omics approaches can generate an unprecedented amount of data, enabling the analysis of highly complex features and providing a general overview of the underlying processes.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. Z and L. M acknowledge co-funding from Next Generation EU, in the context of the National Recovery and Resilience Plan, M4C2 Investment 1.5-ECS00000017, Tuscany Health Ecosystem (THE), CUP B83C22003920001. This resource was co-financed by the Next Generation EU. The views and opinions expressed are only those of the authors and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them. V. V. acknowledges her gratitude to PRIN Prot. 2022JMFC3X.References
- P. Chandrangsu, C. Rensing and J. D. Helmann, Nat. Rev. Microbiol., 2017, 15, 338–350 CrossRef CAS PubMed.
- S. Medici, M. Peana, V. M. Nurchi, J. I. Lachowicz, G. Crisponi and M. A. Zoroddu, Coord. Chem. Rev., 2015, 284, 329–350 CrossRef CAS.
- T. G. Benedek, J. Hist. Med. Allied Sci., 2004, 59, 50–89 CrossRef PubMed.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- S. Singh and R. Sivakumar, J. Infect. Chemother., 2004, 10, 307–315 CrossRef PubMed.
- C. F. Shaw, Chem. Rev., 1999, 99, 2589–2600 CrossRef CAS PubMed.
- C. Manyi-Loh, S. Mamphweli, E. Meyer and A. Okoh, Molecules, 2018, 23, 795 CrossRef PubMed.
- J. O'Neill, Tackling Drug-Resistant Infections Globally: Final Repory and Recommendations, Review on Antimicrobial Resistance, 2016 Search PubMed.
- K. D. Mjos and C. Orvig, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS PubMed.
- D. J. Barillo, A. R. Barillo, S. Korn, K. Lam and P. S. Attar, Burns, 2017, 43, 1189–1194 CrossRef PubMed.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. H. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Chem. Sci., 2020, 11, 2627–2639 RSC.
- A. Frei, Antibiotics, 2020, 9, 90 CrossRef CAS PubMed.
- C. N. Morrison, K. E. Prosser, R. W. Stokes, A. Cordes, N. Metzler-Nolte and S. M. Cohen, Chem. Sci., 2020, 11, 1216–1225 RSC.
- Z. ChunYan, Y. RuJian, W. LiQiang, H. HaiYan, W. JinTao, L. XiangWen, D. XueMin and X. YanShi, Eur. J. Med. Chem., 2022, 240, 114562 CrossRef PubMed.
- R. Wang, M. Wei, X. Wang, Y. Chen, Y. Xiong, J. Cheng, Y. Tan, X. Liao and J. Wang, J. Inorg. Biochem., 2022, 236, 111954 CrossRef CAS PubMed.
- V. C. Nolan, L. Rafols, J. Harrison, J. J. Soldevila-Barreda, M. Crosatti, N. J. Garton, M. Wegrzyn, D. L. Timms, C. C. Seaton, H. Sendron, M. Azmanova, N. P. E. Barry, A. Pitto-Barry and J. A. G. Cox, Curr. Res. Microb. Sci., 2022, 3, 100099 CAS.
- G. Gasser and N. Metzler-Nolte, Curr. Opin. Chem. Biol., 2012, 16, 84–91 CrossRef CAS PubMed.
- T. Gianferrara, I. Bratsos and E. Alessio, Dalton Trans., 2009, 7588 RSC.
- J. E. Waters, L. Stevens-Cullinane, L. Siebenmann and J. Hess, Curr. Opin. Microbiol., 2023, 75, 102347 CrossRef CAS PubMed.
- F. Li, F. Liu, K. Huang and S. Yang, Front. Bioeng. Biotechnol., 2022, 10, 827960 CrossRef PubMed.
- A. M. Pitz, G. W. Park, D. Lee, Y. L. Boissy and J. Vinjé, Gut Microbes, 2015, 6, 93–100 CrossRef CAS PubMed.
- C. Roder and M. J. Thomson, Drugs R&D, 2015, 15, 13–20 CrossRef CAS PubMed.
- H. P. Varbanov, F. Kuttler, D. Banfi, G. Turcatti and P. J. Dyson, PLoS One, 2017, 12, e0171052 CrossRef PubMed.
- M. I. Cassetta, T. Marzo, S. Fallani, A. Novelli and L. Messori, BioMetals, 2014, 27, 787–791 CrossRef CAS PubMed.
- S. Thangamani, H. Mohammad, M. F. N. Abushahba, T. J. P. Sobreira, V. E. Hedrick, L. N. Paul and M. N. Seleem, Sci. Rep., 2016, 6, 22571 CrossRef CAS PubMed.
- P. She, Y. Liu, Y. Wang, F. Tan, Z. Luo and Y. Wu, J. Appl. Microbiol., 2020, 128, 88–101 CrossRef CAS PubMed.
- Y. Yu, H. Zhao, J. Lin, Z. Li, G. Tian, Y. Y. Yang, P. Yuan and X. Ding, Int. J. Antimicrob. Agents, 2022, 59, 106582 CrossRef CAS PubMed.
- H. Tsubery, I. Ofek, S. Cohen and M. Fridkin, J. Med. Chem., 2000, 43, 3085–3092 CrossRef CAS PubMed.
- P. She, L. Zhou, S. Li, Y. Liu, L. Xu, L. Chen, Z. Luo and Y. Wu, Front. Microbiol., 2019, 10, 2453 CrossRef PubMed.
- D. V. Zurawski and M. K. McLendon, Antibiotics, 2020, 9, 155 CrossRef CAS PubMed.
- S. Dwarakanath, J. G. Bruno, T. N. Athmaram, G. Bali, D. Vattem and P. Rao, Folia Microbiol., 2007, 52, 31–34 CrossRef CAS PubMed.
- L. Yu, Z. Shang, Q. Jin, S. Y. Chan, W. Hong, N. Li and P. Li, Adv. Healthcare Mater., 2023, 12, 2202207 CrossRef CAS PubMed.
- A. Frei, A. D. Verderosa, A. G. Elliott, J. Zuegg and M. A. T. Blaskovich, Nat. Rev. Chem, 2023, 7, 202–224 CrossRef CAS PubMed.
- K. Mezgebe and E. Mulugeta, Med. Chem. Res., 2024, 33, 439–463 CrossRef CAS.
- Z. Zong, G. Tian, J. Wang, C. Fan, F. Yang and F. Guo, Pharmaceutics, 2022, 14, 2790 CrossRef CAS PubMed.
- P. Pachori, R. Gothalwal and P. Gandhi, Genes Dis., 2019, 6, 109–119 CrossRef PubMed.
- A. Beceiro, M. Tomás and G. Bou, Clin. Microbiol. Rev., 2013, 26, 185–230 CrossRef CAS PubMed.
- M. Claudel, J. V. Schwarte and K. M. Fromm, Chemistry, 2020, 2, 849–899 CrossRef CAS.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |