Open Access Article
Open Access ArticleAn ensemble model of machine learning regression techniques and color spaces integrated with a color sensor: application to color-changing biochemical assays†
Min Joha,
Surjith Kumarana,
Younseo Shina,
Hyunji Chaa,
Euna Oha,
Kyu Hyoung Lee b and
Hyo-Jick Choi
b and
Hyo-Jick Choi *a
*a
aDepartment of Chemical and Materials Engineering, University of Alberta, Edmonton, AB T6G 1H9, Canada. E-mail: hyojick@ualberta.ca
bDepartment of Materials Science and Engineering, Yonsei University, Seoul 03722, Republic of Korea
First published on 20th January 2025
Abstract
Non-destructive color sensors are widely applied for rapid analysis of various biological and healthcare point-of-care applications. However, existing red, green, blue (RGB)-based color sensor systems, relying on the conversion to human-perceptible color spaces like hue, saturation, lightness (HSL), hue, saturation, value (HSV), as well as cyan, magenta, yellow, key (CMYK) and the CIE L*a*b* (CIELAB) exhibit limitations compared to spectroscopic methods. The integration of machine learning (ML) techniques presents an opportunity to enhance data analysis and interpretation, enabling insights discovery, prediction, process automation, and decision-making. In this study, we utilized four different regression models integrated with an RGB sensor for colorimetric analysis. Colorimetric protein concentration assays, such as the bicinchoninic acid (BCA) assay and the Bradford assay, were chosen as model studies to evaluate the performance of the ML-based color sensor. Leveraging regression models, the sensor effectively interprets and processes color data, facilitating precision color detection and analysis. Furthermore, the incorporation of diverse color spaces enhances the sensor's adaptability to various color perception models, promising precise measurement, and analysis capabilities for a range of applications.
Introduction
The emergence of novel technologies has led to a significant surge in research efforts focused on sensors, specifically aimed at enhancing biological assays and quantifying various biological components, such as proteins, cells, or pathogens.1 One of the fundamental principles governing device operation involves the utilization of sensors employing color changes, widely employed across various fields. This approach is favored due to its inherent advantages, allowing for both qualitative prediction and quantitative analysis of color alterations. Colors are visual perceptions shaped by the interaction of visible light and its distribution of wavelengths, constituting a central concept in optics.2 Their color differentiation serves as physical indicators in various applications, including water quality tests, explosive tests, explosive tests, pH, glucose, starch sensors, drug tests, humidity measurement, and vital diagnostic indicators in biological and chemical assays.3–6 Traditionally, photoelectric and visual colorimetry were used to measure concentration by observing color changes with a spectrophotometer or with the naked eye.7Biologists and chemists often utilize color to track reactions of interest. However, due to the lower accuracy and precision of visual colorimetry, instruments like spectrophotometer are typically needed for quantitative data. Spectrophotometers provide more accurate color change resolution and are preferred for concentration determination applications.8 These colorimetric tests use spectroscopic absorbance measurements and a calibration curve to determine analyte concentration accurately.9 However, visual colorimetry methods encounter significant operational limitations on-site, including user interpretation errors and environmental inconsistencies, leading to unreliable outcomes. Currently, digital cameras, smartphones, and scanners, can be used for image capturing, but their accuracy of color information is affected by ambient light changes and built-in automatic image correction.10,11 Traditional human visual assessment is limited, as the human eye can struggle to discern subtle changes accurately and consistently, especially when the changes indicate the presence of small amounts of an analyte. Human error, combined with external factors like lighting, temperatures, and sample distance, complicates the reliability of these readings. Converting these color changes into quantitative data is challenging due to subjective and error-prone quantifiable color indices.12
Using controller boards is deemed a simpler alternative for building automated systems for rapid execution.13 Color sensors offer greater quality, portability, do-it-yourself (DIY) capabilities, and cost-effectiveness compared to spectrophotometers, encouraging their adoption over more expensive spectrometers.14 In many colorimetric procedures, color information is often described using color spaces like red, green, blue (RGB).15 However, RGB has been found to be complex in terms of human perception.16 RGB lacks perceptual uniformity and intuitive control over hue, saturation, and brightness. Additionally, since RGB values depend on device characteristics, colors displayed on various devices can differ, even when using identical RGB values. Accurate readings and reproducible assessment of colors, both qualitatively and quantitatively, are crucial.17 Perceptually uniform spaces like cyan, magenta, yellow, key (CMYK), CIELAB (L*a*b*) and approximately uniform spaces such as hue, saturation, lightness (HSL) and hue, saturation, value (HSV) offer more intuitive color representation, where measured differences reflect human perception. HSV and HSL separate hue from intensity, aiding in color recognition, while CIELAB provides high accuracy for colorimetric analysis. Combining RGB's practicality with the perceptual advantages of other color spaces ensures efficient and accurate color quantification. Colorimetric sensors have individual sensors for red, green, and blue, each detecting colors specific light wavelengths. These sensors operate within a frequency range of 2 Hz to 500 kHz and convert the detected values into a scale from 0 to 255. By merging these values, the appropriate color code can be obtained.18 To mitigate uncertainties in human vision, digital colorimetry requires image calibration algorithms for the visible color space. Converting the RGB system to human-perceptible color spaces like HSL or HSV addresses this limitation by ensuring linear changes in chroma or color intensity.19 Additionally, the implementation of machine learning (ML) techniques can enhance data analysis and interpretation by uncovering insights, making predictions, automating processes, and facilitating decision-making, especially with non-linear data.20 It has been reported that ML algorithms excel in classifying, discriminating, and predicting unknown samples by uncovering latent patterns within voluminous, noisy, or intricate datasets.21 Thus, by leveraging ML approaches, colorimetric sensor devices have been devised to offer competitive accuracy, low-cost, convenient, non-destructive methods, and enhanced colorimetric assays for chemical and biological applications. The advantages of ML algorithms include adaptability to different settings, making them applicable to sensors for real-time analysis.22,23 Despite advancements using ML technologies, color sensors still fall short of human color processing capabilities. Key drawbacks include slow speed, low identification efficiency, poor real-time performance, and limited regression models. Hence, achieving optimal performance in color sensor devices requires prioritizing precise color recognition, as the accuracy and reliability of their data depend on it.
In this work, we used an ML algorithm to replicate the human ability to recognize patterns and applied it to various color models, some based on human perception, including RGB, HSL, and HSV, as well as the CMYK, and CIELAB. Considering the closer alignment of the HSL model with human perception, it is logical to explore whether ML algorithms could benefit from adopting this color model. By utilizing the HSL model, the fabricated color sensor device takes an intuitive, human-aligned approach to identifying subtle color changes. To demonstrate the effectiveness of the HSL model in detecting saturation and hue differences, it was tested to predict protein concentration accurately. To this end, conventional protein assays such as the bicinchoninic acid (BCA) and Bradford assays were compared with image-based colorimetric measurement in the HSL color space using ML algorithms. Currently, widely employed colorimetric protein assay techniques require spectrophotometers, limiting their versatility. However, image-based colorimetric detection has emerged as a cost-effective alternative for field applications compared to traditional methods such as spectrophotometry, colorimetry, and fluorometry.24 Recognized for its ability to perform both qualitative and quantitative protein analysis, this method is highly considered one of the most promising approaches in protein assays. Given their biological significance, accurate methods for detecting, identifying, and quantifying proteins are routinely employed for diagnostic purposes in clinical settings, including proteomics, UV-vis spectrometry, electrophoresis, and immunoblotting.25 Paving the way for advancements in ML, this approach holds the potential to extend beyond the conventional RGB model, aligning more closely with human perception and interpretation of color. ML models offer a significant advantage in image-based colorimetric detection, resilience against unwanted variations.26 Current methods for detecting color changes often rely on identifying a single type of change using regression models like linear or logistic regression. In this study, we utilize four machine learning models — random forest regressor (RFR), gradient boosting regressor (GBR), support vector regressor (SVR), and multi-layer perceptron (MLP) — tailored for different datasets, with the aim of enhancing prediction accuracy by capturing linear correlations between dependent variables. MLPs, a class of artificial neural networks (ANN), have been utilized to model the CIELAB color space due to their capacity to manage non-linear relationships and complex interactions.27 RFR combines decision trees for high accuracy but can be resource intensive, achieved over 90% accuracy in predicting peroxide.28 GBR sequentially corrects errors, offering high accuracy for complex patterns with prediction errors of 10–20% in dye concentration estimation.29 SVR excels at modeling nonlinear relationships in color spaces like CIELAB, achieving mean absolute percentage error (MAPE) below 12% and low RMSE for chromium(VI) and iron(III).30 MLPs use deep neural layers to model complex relationships, delivering near-human precision in color reconstruction and outperforming traditional methods.31 Thus, multiple machine learning models help reduce prediction error, while lower ensemble complexity aids in reducing computational demands.32 Additionally, the ML framework introduces an end-to-end processing pipeline for color detection using non-RGB sensing devices such as HSL, HSV, CMYK, and CIELAB, enabling rapid detection and adaptation to diverse colorimetric applications, including detecting chemical analytes and biological assays.
Experimental section
Fabrication of a raspberry pi-based colorimetric sensor
The micro-BCA protein assay kit and Bradford assay reagent were purchased from Thermo Fisher Scientific (Waltham, IL, USA) and Bio-Rad (Hercules, CA). The absorbance was measured using a Bio-Rad iMark Microplate reader (Hercules, CA, USA). Raspberry Pi 4B with a TCS3200 color sensor (DFRobot) was used for color sensing, measuring RGB frequencies through color filters and outputting RGB values, predicted results, and measurement indices. The sensor, operated by TCS3200.py, reads color frequencies and outputs RGB values. The system included a Raspberry Pi 4 Model B, a 1602 LCD display with an I2C chip, and a power supply. The I2C chip, purchased from Amazon (Toronto, ON, Canada), displays RGB numbers and measurement indices as defined in I2CLCD.py. Components were arranged according to the pin/port configuration shown in Fig. S1.† The TCS3200 sensor, emitting LED light and capturing reflected light, accurately read RGB values of colors.The schematic of the Raspberry Pi-connected RGB device and color palette for BCA and Bradford assays is in Fig. S1.† The sensor's response to light was evaluated by fixing an LED light source to a 3D-printed support for the 96-well plate, ensuring direct radiation onto the sensor. The RGB output values from the sensor, derived from colored albumin solutions, were used to develop an algorithm for reporting RGB sensor outputs. Subsequently, coding tasks were performed, and the generated code was applied to the device. The resulting data was then compared with spectrophotometry readings for validation.
Bicinchoninic acid (BCA) and Bradford protein assays
The Biuret reaction, BCA assay, and Bradford assay have been commonly used in estimating protein content in a solution by measuring absorbance.33–35 In this work, protein concentration was determined using a micro-BCA and Bradford protein assays, according to the manufacturer's protocols using bovine serum albumin (BSA) as reference standards. Briefly, for micro-BCA assay, 100 μL of each standard replicate is pipetted into their respective spots on the labeled 96 well plate (Greiner Bio-One, Kremsmünster, Austria), followed by the addition of 100 μL of the working reagent. After shaking for 30 seconds, the plate is then covered with a lid and incubated at 60 °C for 1 hour. Afterwards, the loaded standards and samples are measured for absorbance at 560 nm using a plate reader (iMark, Bio-rad, Hercules, CA) followed by subtracting the average absorbance reading of the blank standard replicates from those of all other standard sample replicates. The BCA analysis quantifies proteins by converting Cu2+ to Cu+ through the bicinchoninic acid (BCA) complex in an alkaline solution. This process involves two key reactions. First, at low temperatures (37 °C), BCA interacts with copper ions and protein residues, where the protein-copper ion complex reduces Cu2+ to Cu+ in an alkaline medium (the biuret reaction). Second, at higher temperatures (60 °C), an intense purple color develops due to the interaction of the reduced Cu+ complex with two bicinchoninic acid (BCA) molecules. The schematic mechanism of the BCA assay for protein quantification is provided in the ESI (see Fig. S2†).For the Bradford assay, 50 μL of standard containing BSA was prepared and placed into a 96-well plate. Then, 150 μL of Bradford assay reagent (Bio-Rad, Hercules, CA) was added to the protein standard and incubated for 10 minutes at room temperature. The absorbance was measured at 595 nm using a plate reader. The Bradford Assay mechanism relies on the binding of Coomassie Brilliant Blue G-250 dye to proteins. The dye (465 nm) interacts with proteins through hydrophobic interactions (involving residues such as phenylalanine and tryptophan) and electrostatic interactions specifically, the sulfonic group of the dye and positively charged guanidino or arginine groups. Upon protein binding, the protein-dye complex causes a shift in the dye's absorption maximum from 465 nm to 595 nm. The intensity of the absorption at 595 nm is directly proportional to the protein concentration in the solution, making it a reliable quantitative method for protein quantification. The schematic Bradford assay mechanism is shown in Fig. S3.†
Color sensor calibration and ML analysis on raspberry Pi
The Raspberry Pi 4B (1.2 GHz 64 bit quad-core ARM Cortex-A53 CPU, 1 GB RAM, 40 GPIO pins) was used to perform ML tasks for collecting RGB frequencies (see Fig. S4†). Python programs were developed on a local laptop for calibration, light intensity, and RGB frequency detection. The color sensor, utilizing a 1602 LCD display with an I2C chip and a TCS3200 sensor, is controlled by I2CLCD.py and TCS3200.py. The TCS3200 sensor measures the color, and the obtained raw RGB frequency readings are first normalized using min–max scaling to correct for light intensity variations across the plate. This normalization is part of a base length correction process, which begins with calculating the sum of RGB frequency values for each well filled with DI water. The average RGB sum is then determined from the sample rows, and the row with the highest average RGB sum is identified as the brightest row and assigned 100% brightness. The obtained outputs data are in CSV format, displaying RGB frequencies, RGB numbers, predicted values, and measurement indices. The TCS3200 sensor reads RGB light intensity through an 8 × 8 photodiode matrix, dividing 64 photodiodes into four color groups. Calibrating the sensor was necessary to obtain accurate RGB numbers. Calibration was performed using python code to compile RGB frequency values, setting liquid white to (255, 255, 255) and liquid black to (0, 0, 0). Each RGB value was then scaled to ensure accurate readings based on a calibration object. The calibration took place on a 96-well plate placed on a lightbox emitting diffused white light (8500 K). The Python script calibration.py generated sensor calibration data, with the sensor, capped by a 3D-printed cover, placed on the samples to capture color readings. The program recorded 10 readings, and the minimum and maximum frequencies for black and white were stored in calibration.txt corresponding to RGB (0, 0, 0) and RGB (255, 255, 255). The sensor was positioned 1.7 cm above the sample meniscus, recording 10 readings, with frequencies stored in calibration.txt. To read experimental samples, BCA and Bradford protein assays were prepared in a 96-well plate on the lightbox. The read_color.py program was run to estimate protein concentration. It returns outputs in the RGB frequency format as a CSV file and displays various measurements such as RGB numbers, predicted values, and measurement index. An I2C chip in raspberry pi address the functions to display texts (machine status) and numbers (RGB frequency) are defined in I2CLCD.py.Color analyses involved a database of protein concentrations using 216 BCA assay samples (0 to 200 μg mL−1). Results were evaluated with calibration curves (see Fig. S5†). The TCS3200 sensor collected RGB frequencies of samples with known and unknown protein concentrations, automatically stored in a database. To build the regression models, we utilized a set of machine learning algorithms form the scikit-learn library. Specifically, we constructed four models: such as RFR, GBR, SVR, and MLP. These models were organized into a Python dictionary to facilitate efficient management, model comparison, and iteration during training. Through the interactions of various parameters of the four machine learning models, it automatically finds the best parameter set to use for each model. Using 20% of the dataset as test samples for each protein concentration measurement, the program returns metric scores, which include mean absolute error (MAE), mean squared error (MSE), root mean squared error (RMSE), and coefficient of determination (R2) scores, to interpret the results and select the best model. The program also illustrates prediction and residual plots to visualize the results.
Results
Design of software architecture and operation of color sensor
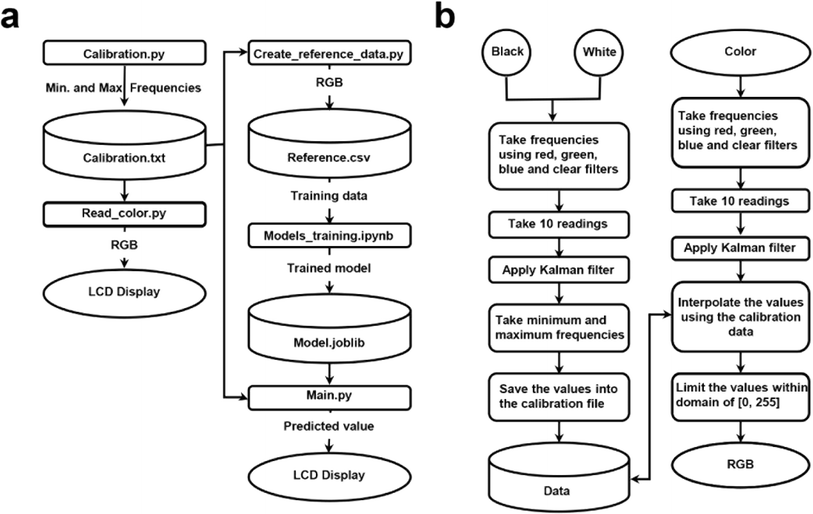
Fig. 1 illustrates the decision sequence for calibration and the software architecture, highlighting data management and operational procedures. In Fig. 1a, the calibration procedure is initiated by users, triggering the execution of the calibration.py script this script acquires and records the minimum and maximum frequencies of the red, green, and blue color channels, corresponding to black [0, 0, 0] and white [255, 255, 255] in RGB format. The read_color.py script then reads the color using the calibrated data from calibration.txt, and the RGB values obtained are then displayed on an LCD screen. Color images saved in the RGB format exhibit unevenness, posing challenges in assessing color similarity based on their proximity in the RGB color space.33 Due to sensor's sensitivity to light variations, its measurements can be affected by variability arising from calibration disparities caused by environmental conditions. Therefore, calibration for each operation is essential to account for different ambient lighting, material states or surface finishes, and other factors.36,37 This ensures accurate color measurement and reduces data variability. Our approach of using ambient light from a lightbox and specific frequency scaling settings mitigates the impact of external light conditions and aligns with the main programs. During the calibration and color measurement processes using BCA and Bradford assays, the dedicated LED is not used to avoid external interference. Instead, ambient light from a lightbox (white light, power density: 2 mW cm−2) is harnessed, with frequency scaling set to operate at 20%. The create_reference_data.py script generates reference data in RGB format, which is saved in reference.csv. This reference data is used to train a regression model within the models_training.ipynb notebook, resulting in a trained model saved as model.joblib. The main.py script uses this trained model to predict color values.In Fig. 1b, frequencies for both black and white and color samples are measured using red, green, blue, and clear filters. A Kalman filter is used to improve frequency reading accuracy and reduce noise by consolidating ten post-calibration measurements into a single frequency reading, a technique commonly used in precise measurements like GPS or telecommunications.26 Five raw RGB readings are averaged during data creation, ensuring deviations between consecutive readings remain below three units for each color channel, thereby refining the precision of the RGB readings. Also, this method's real-time computational complexity (input data vs. number of operations) is efficient for mobile applications due to the matrix inversion's cubic Big O complexity (n3), where n is the state vector dimension.
The embedded color reading function converts raw frequency reading into RGB values, scaling them between 0 and 255. Frequencies below the minimum are set to 0, and those above the maximum are set to 255, with intermediate values interpolated. This ensures precis color data representation. Fig. S6a† shows a linear relationship between the sum of RGB frequencies and numbers, confirming sensor reliability. Fig. S6b† illustrates the ratio of individual colors (R, G, B) to the sum of RGB with increasing protein concentration, highlighting the sensitivity of the colorimetric method to changes in protein levels.
Data was collected and saved in five color spaces (RGB, HSL, HSV, CMYK, and CIELAB) for ML procedures. Four regression models – RFR, GBR, SVR, and MLP – were trained using optimal parameters and evaluated on a test dataset.38,39 Regression models deliver accurate color predictions by reducing color calibration errors and are suitable for real-time application on devices with limited computational resources.40,41 Those models are chosen for color sensing devices due to their efficiency in handling continuous data outputs, which is crucial for capturing subtle differences in color shades.
Variations of colorimetry models and their analysis
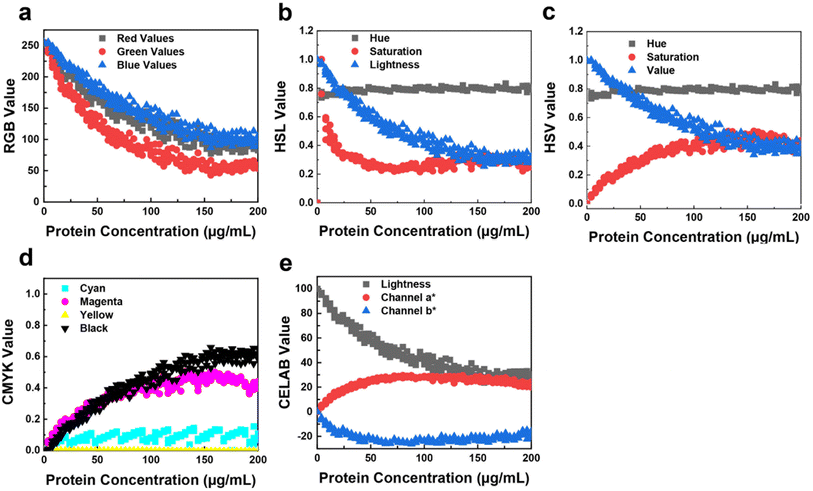
Utilizing a multivariate calibration method, this study maximized covariance between color values and protein concentration. A training matrix was developed with 50 samples of BSA concentrations ranging from 0 to 200 μg mL−1, prepared in quintuplicate. Each model provides a scattered plot showing the trend across each color channel (Fig. 2). The scatter plot for the RGB model (Fig. 2a) reveals a non-linear, exponential decay trend across each color channel, a common occurrence in coloring assays due to the coexistence of red, green, and blue forms as protein concentration increases.19When transitioning to other colorimetry models, distinct trends emerge: minimal hue variations, increased saturation, and decreased lightness, aligning with expected outcomes. The relationship between RGB values and protein concentration is crucial in Bradford colorimetric analysis, affecting color intensity and distribution. The HSV color space is employed instead of RGB for better detection accuracy, as it separates light effects from color information.
The regression model was tested under ambient light conditions during the BCA assay (Fig. 2). As protein concentration increased, RGB values decreased, resulting in a darker blue color (Fig. 2a). In the HSL model, hue remained consistent while saturation increased and lightness decreased, indicating a darker color (Fig. 2b). Similarly, the HSV model showed consistent hue with increased saturation and decreased value (Fig. 2c). In Fig. 2d, the CMYK model showed consistent cyan with increasing magenta, yellow, and black values, resulting in a more intense purple (Fig. S6c†). Finally, Fig. 2e demonstrates that in the CIELAB model, lightness decreases, a* shifts positively, and b* shifts negatively, indicating a transition to a darker blue with increasing protein concentration (Fig. S7a†). These results compare the protein concentration-dependent RGB components, showing significant variance due to uneven ambient lighting. In the HSL and HSV models (Fig. S7b and c†), the hue shows minimal variation, while saturation increases and lightness decreases, as expected. Periodic dimming can introduce variances in ML predictions, especially at higher protein concentrations, where subtle color shifts cause trend divergence. Additionally, the transparency of the 96-well plate may lead to minor reading inaccuracies due to the influence of neighboring colors.
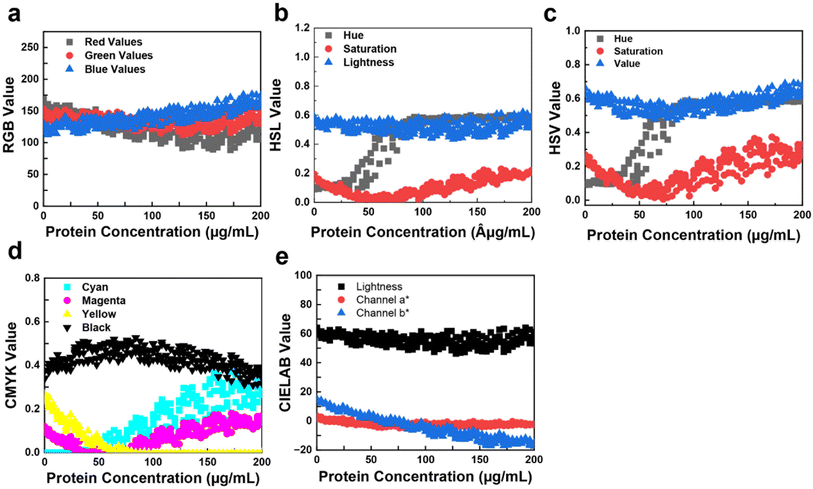
The relationship between RGB values (Fig. S8a†) and protein concentration is crucial for Bradford colorimetric analysis (Fig. 3). As protein concentration increases, changes occur in the intensity and distribution of colors captured by RGB, HSL, HSV, CMYK, and CIELAB sensors. The HSV model is preferred for its better detection accuracy in image analysis as it separate light effects from color information.
In Fig. 3a, as protein concentration increases, red decreases and blue increases, indicating a color shift from yellow to blue (Fig. S8b†). Fig. 3b shows HSL values vs. protein concentration, where hue sharply increases, indicating a yellow-to-blue shift, with saturation decreasing and then intensifying blue around 50 μg mL−1. This pattern is reflected in the HSV graph (Fig. 3c), where hue, lightness, and value increase around 50 μg mL−1 (Fig. S7a†), indicating a color shift. The CMYK values vs. protein concentration (Fig. 3d) show cyan increasing from 50 μg mL−1 (Fig. S9b†), magenta first decreasing then increasing, and yellow decreasing and stabilizing around 60 μg mL−1, indicating the yellow-to-blue transition. Finally, Fig. 3e shows CIELAB vs. protein concentration, where the b* channel decreases, signifying a transition from yellow to blue.
Comparing various color models, periodic dimming is noticeable in the RGB readings of the Bradford assay dataset (Fig. 3). Lower protein concentrations show a distinctive brown hue (higher red, lower blue values), shifting to a predominant blue shade (higher blue, lower red values) at higher concentrations. Notably, the a* channel of CIELAB exhibits a consistent trend ranging approximately from 10 to −10 (Fig. S9c†), potentially enhancing model performance. Similar to the BCA assay results, divergences in trends become more pronounced beyond approximately 200 μg mL−1, likely due to subtle variations in blue color intensity in the Bradford assay.
Evaluation of the four regression models across five colorimetry models
Various supervised ML algorithms were applied to an experimentally established dataset measuring protein solution concentrations. The initial hypothesis favored HSL or HSV models for superior performance, given the expected variability in hue, saturation, or lightness values typical in biological color determinations.During the BCA assay, the sensor captured color readings ranging from 0 to 200 μg mL−1 in 4 μg mL−1 increments. The dataset included 204 measurements from four 96-sample trays, each containing 51 samples. RGB values showed periodic dimming occurrences nine times across the 96-tray setup, indicating inconsistent ambient lighting affecting each row. However, a clear trend emerged with increasing protein concentration: all three RGB values decreased. This decline led to higher saturation levels in HSV and HSL, increased magenta and black values in CMYK, and reduced lightness in CIELAB, HSL, and HSV systems.
Notably, the consistent rises in HSV saturation or CMYK magenta are expected to influence effective model training. Periodic dimming introduces prediction variances when identifying influential features for ML. The two predominant trends begin to diverge at higher protein concentrations, likely due to subtle color shifts at elevated levels. Additionally, the transparency of the 96-well tray may cause minor reading inaccuracies influenced by neighboring colors. Regression models including MLP, GBR, SVR, and RFR were trained on the dataset shown in Fig. 4. Evaluating these algorithms on the test dataset revealed strong performance for protein volumes ranging from 0 μL up to approximately 70 μL. Particularly in HSL and HSV color metrics, predictions closely matched actual values. However, for protein volumes exceeding 70 μL, result consistency diminished, likely due to factors such as high variability in the BCA assay, occasional quantification of different proteins with identical concentrations, and sensitivity to substances like salt, detergents, and reducing agents.42–44 It's noteworthy that the neural network model struggled and did not provide accurate predictions within this higher range, possibly due to nuanced color changes occurring with increasing protein volumes.
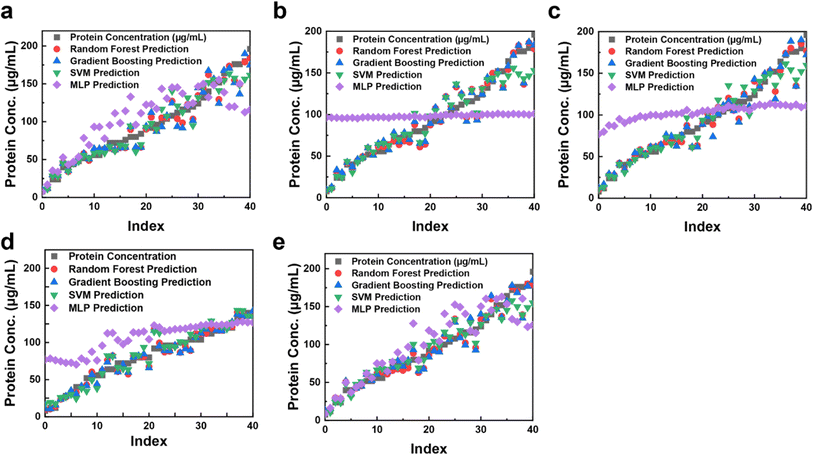 | ||
| Fig. 4 Scatter plots of the predicted protein concentrations from regression models (RFR, GBR, SVR, and MLP) in five different color systems, RGB (a); HSL (b), HSV (c), CYMK (d), and CIELAB (e). | ||
The RGB, HSL, HSV, CMYK, and CIELAB responses were plotted using various regression models to predict the hue coordinate for the mixed indicator, ranging between 0 and 45 (Fig. 4). This broad range indicates these color coordinates provide high-resolution measurements suitable for diverse regression models. Experimental data were fitted with a fourth-order polynomial curve, enabling concentration determination of unknown solutions based on RGB, HSL, HSV, CMYK, and CIELAB values obtained from the device.
Fig. 4a illustrates protein prediction using the RGB model with various regression techniques. Regression models trained on assay data achieved accurate predictions of protein concentrations. During cross-validation, RFR, GBR, and SVR demonstrated optimal performance within the RGB model, while HSL, HSV, CMYK, and CIELAB (Fig. 4b–e) also showed high performance. However, MLP deviated in fitting compared to other models. MLP, a nonlinear neural network, captures complex relationships due to its layers and activation functions but requires extensive data for effective training. Performance variations may arise from its flexibility, sensitivity to hyperparameters, and differences in feature scaling and data interpretation. These results highlight the color sensor's potential for accurately predicting protein concentrations. The performance across different color models and ML techniques underscores their applicability in various color-to-concentration applications. The TCS3200 color sensor detects red, green, blue, and overall light using respective filters, influenced by factors like ambient color temperature, reflections, surface colors, finishes, and sensor angle relative to light source. While these factors minimally alter hue values, they noticeably affect saturation and lightness values, emphasizing the need for controlled lighting systems to enhance dataset quality. Addressing signal nonlinearity within sensor devices is critical to minimizing measurement errors, often addressed through artificial neural network models.21 Furthermore, ensuring linearity between observed and expected protein concentrations enhances the reliability of bioanalytical approaches.
Data analysis of the colorimetric assay
Evaluating the regression model (Fig. 5) is critical to understanding its predictive performance for unknown sample concentrations. The model's effectiveness is determined by comparing its predicted outcomes to the actual values, aiming to minimize prediction errors for optimal results. Common metrics used in regression models include MAE, MSE, RMSE, and the R2 score.45,46 These metrics assess the model's accuracy and reliability in predicting protein concentrations. To evaluate the accuracy of the models, the statistical parameters mean absolute error (MAE) (eqn (1)), mean square error (MSE) (eqn (2)), root mean squared error (RMSE) (eqn (3)), and correlation coefficient (R2) (eqn (4)) were employed.
 | (1) |
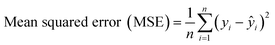 | (2) |
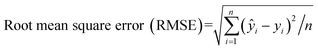 | (3) |
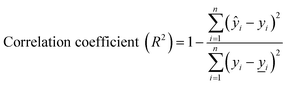 | (4) |
![[y with combining low line]](https://www.rsc.org/images/entities/i_char_0079_0332.gif) i is the average of the actual cumulative confirmed cases.
i is the average of the actual cumulative confirmed cases.
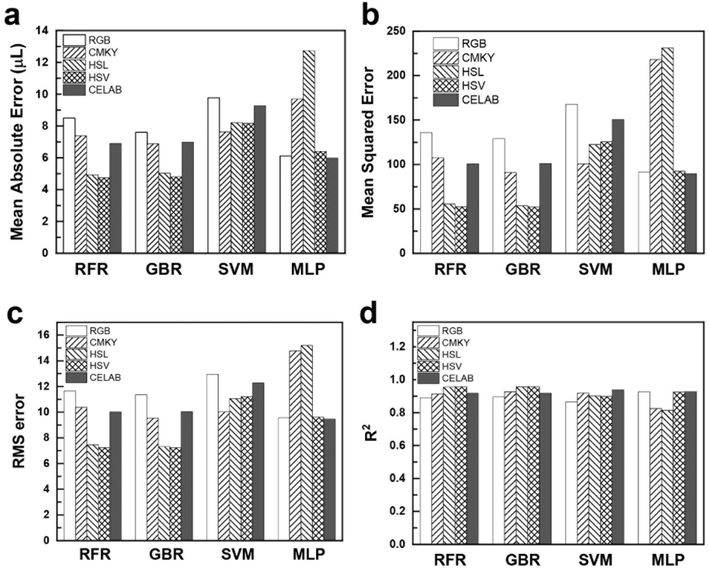
In protein concentration assays, RFR and GBR consistently outperform other models based on metric scores. For instance, using the RGB colorimetry model (Fig. 5a), RFR achieves an MAE of 8.1, GBR records 8.95, and SVR achieves 8.21. In comparison, MLP shows a higher MAE of 11.02. This highlights the superior performance of tree-based models like RFR and GBR in accurately detecting protein concentrations, particularly in capturing subtle color differences and intricate dataset patterns. In terms of MSE (Fig. 5b), tree-based models demonstrate exceptional accuracy with the HSL and HSV color models. For HSL, RFR and GBR achieve MSE values of 93.1 and 96.65, respectively. In the HSV model, RFR and GBR record MSE values of 85.35 and 102.49, respectively. These errors consistently remain below ±5, indicating robust performance. Similarly, the RMS error graph (Fig. 5c) reveals the lowest errors for HSL and HSV using RFR (9.64 for HSL and 9.23 for HSV) and GBR (9.83 for HSL and 10.12 for HSV). In contrast, MLP shows the highest errors (31.32 for HSV and 35.49 for HSL). The R2 score (Fig. 5d) further confirms the models' accuracy, with RFR and GBR both achieving a score of 0.96 in the HSV model, indicating that these models explain over 95% of the variance in the dataset. SVR also demonstrates strong performance, particularly with the CMYK color model, achieving an MAE of 9.51, closely comparable to RFR's 9.54. This underscores SVR's effectiveness in capturing inherent patterns in the CMYK color space. Conversely, MLP excels in the CIELAB model with an R2 of 0.96, the best among all models for this color system, highlighting the neural network's capability. Detailed error evaluations are provided in ESI Table S1.†
In our study, we used 20% of the input datasets to generate performance metrics. While increasing data volume can enhance accuracy, it also demands more resources and time. Our goal was to achieve robust performance indicators with minimal number of features. We employed a combination of MLP and various color spaces to optimize protein concentration predictions, ensuring adaptability across diverse datasets. Prioritizing regression models over classifiers allowed us to achieve precise quantification of protein concentrations. Unlike existing devices relying on a single standard concentration, our method considers a wide range of concentrations using ML techniques. Our approach offers improved detection accuracy, distinguishing our color sensor from traditional absorbance-based analyzers.
The color sensor device facilitates colorimetric-based BCA and Bradford assays, providing direct protein quantification crucial for assessing protein levels in biological samples. This can significantly contribute to disease diagnosis, monitoring, and treatment. Our sensor measures RGB signals from the BCA assay plate, demonstrating its potential to replace multiplexed analyses such as commercial microplate readers (see Table S2† for the comparison with previous work).
To assess the colorimetric sensor's performance, we conducted an evaluation using the BCA protein assay and compared protein estimation across multiple models. The sensor detected BSA concentrations ranging from 0 to 160 μg mL−1 during BCA assays and captured RGB frequencies in just 10 seconds, faster than traditional plate readers. The integrated machine learning program enabled precise measurement of RGB intensity in the 96-well plate assay, facilitating quantitative protein analysis.
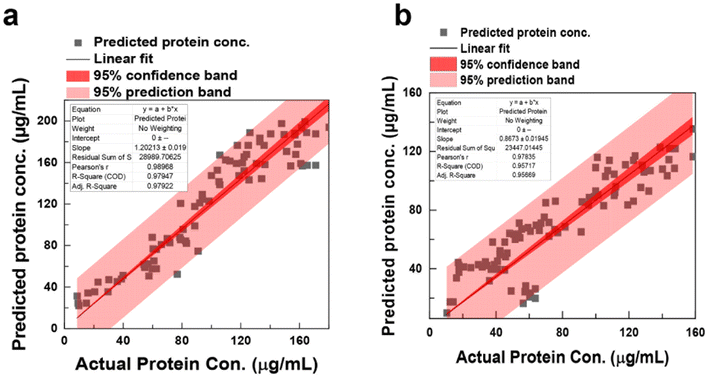
Converting RGB frequency to protein concentration involves a sophisticated ML process. By correlating RGB readings with known protein concentrations, a linear regression curve is established, allowing protein concentration estimation from RGB values. The software's ML algorithms ensure precise RGB intensity measurements for each well in the 96-well plate assay, generating quantitative data for protein concentration analysis. Accuracy depends on the calibration curve quality and experimental consistency. Cross-validation (hold-out cross-validation) assesses model performance, with Fig. 6 illustrating the relationship between the number of principal components and R2, aiding in selecting the optimal components for accurate predictions. The predicted protein concentration was subsequently used to estimate the protein concentration using the BCA and Bradford assays, spanning a concentration range of 0 to 160 μg mL−1. To determine the linear range of these assays, least-squares linear regression equations were computed based on the experimental data, yielding R2 of 0.989 and 0.957 for the BCA and Bradford assays, respectively. A high R2 indicates effectiveness in fitting the model to the observed data, reflecting how well the model captures the underlying trend. The sensitivity of the assays is determined by the slope of the regression lines, yielding 1.20 ± 0.02 for BCA and 0.8673 ± 0.02 for Bradford. The predicted vs. observed plot (Fig. 6) visually represents the model's accuracy and the range of values covered by the experimental data at a 95% confidence level. Finally, using the color sensor technology, we conducted a BCA assay experiment to determine lysozyme protein concentration (see Fig. S10†). This study demonstrates that the proposed machine learning-based color sensor technology can be broadly applied to predict the concentrations of various proteins and monitor biochemical reactions.
Conclusions
In this study, we developed a low-cost, portable RGB detection system using a colorimetric sensor coupled with ML to predict unknown protein concentrations. The sensor was validated using BCA and Bradford protein assays across a range of protein concentrations, establishing a comprehensive dataset. Various regression models were employed to optimize prediction accuracy by leveraging distinctive dataset characteristics. This approach enabled precise establishment of linear relationships between colorimetric signals and detected protein concentrations, ensuring accurate concentration determination. The integration of ML algorithms for interpreting colorimetric data enhances the sensor's utility as a portable and cost-effective prediction tool for diverse colorimetric assays. Further enhancements, such as expanding the dataset and employing advanced techniques like deep learning, promise to improve accuracy and sensitivity, making these tools invaluable in resource-limited environments.Data availability
The complete code for data processing, model training, and testing is provided in a separate link (https://drive.google.com/drive/folders/1qYh6Iy1xSXlTzDGJAsaXtXPG5ZBfxv2w).Author contributions
M. J. and S. K. performed the experiments, curated the data, and wrote the manuscript; Y. S., H. C., E. O. performed experiments, analyzed the data, and edited manuscript; K. H. L conceived the experiments, provided resources, and edited the manuscript; H.-J. C. conceived the experiments, designed the experiments, analyzed the data, provided resources, supervised the research, and wrote the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The funding for this research was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) through the NSERC Discovery Grant (RGPIN-2018-04314) and the National Research Foundation of Korea funded by the Ministry of Education (NRF-2019R1A6A1A11055660).References
- C. D. Flynn, D. Chang, A. Mahmud, H. Yousefi, J. Das, K. T. Riordan, E. H. Sargent and S. O. Kelley, Biomolecular sensors for advanced physiological monitoring, Nat. Rev. Bioeng., 2023, 1, 560–575 CrossRef CAS PubMed.
- P. F. Gao, G. Lei and C. Z. Huang, Dark-field microscopy: recent advances in accurate analysis and emerging applications, Anal. Chem., 2021, 93, 4707–4726 CrossRef CAS PubMed.
- D. H. Brainard, Color and the cone mosaic, Ann. Rev. Vis. Sci., 2015, 1, 519–546 CrossRef PubMed.
- B. B. Lee, The evolution of concepts of color vision, Neurociencias, 2008, 4, 209 Search PubMed.
- L. Xu, H. Wang, Y. Xu, W. Cui, W. Ni, M. Chen, H. Huang, C. Stewart, L. Li and F. Li, Machine learning-assisted sensor array based on poly (amidoamine)(PAMAM) dendrimers for diagnosing Alzheimer's disease, ACS Sens., 2022, 7, 1315–1322 CrossRef CAS PubMed.
- Y. Luo, X. Xiao, J. Chen, Q. Li and H. Fu, Machine-learning-assisted recognition on bioinspired soft sensor arrays, ACS Nano, 2022, 16, 6734–6743 CrossRef CAS PubMed.
- S. Wu, D. Li, J. Wang, Y. Zhao, S. Dong and X. Wang, Gold nanoparticles dissolution based colorimetric method for highly sensitive detection of organophosphate pesticides, Sens. Actuators, B, 2017, 238, 427–433 CrossRef CAS.
- R. G. Bates, Determination of pH: Theory and Practice, 1964 Search PubMed.
- S.-K. Lee, M. Sheridan and A. Mills, Novel UV-activated colorimetric oxygen indicator, Chem. Mater., 2005, 17, 2744–2751 CrossRef CAS.
- D. C. Christodouleas, A. Nemiroski, A. A. Kumar and G. M. Whitesides, Broadly available imaging devices enable high-quality low-cost photometry, Anal. Chem., 2015, 87, 9170–9178 CrossRef CAS PubMed.
- L. Shen, J. A. Hagen and I. Papautsky, Point-of-care colorimetric detection with a smartphone, Lab Chip, 2012, 12, 4240–4243 RSC.
- Y. Shen, S. Modha, H. Tsutsui and A. Mulchandani, An origami electrical biosensor for multiplexed analyte detection in body fluids, Biosens. Bioelectron., 2021, 171, 112721 CrossRef CAS PubMed.
- R. Mehta, J. Sahni and K. Khanna, Internet of things: vision, applications and challenges, Procedia Comput. Sci., 2018, 132, 1263–1269 CrossRef.
- N. Gous, D. I. Boeras, B. Cheng, J. Takle, B. Cunningham and R. W. Peeling, The impact of digital technologies on point-of-care diagnostics in resource-limited settings, Expert Rev. Mol. Diagn., 2018, 18, 385–397 CrossRef CAS PubMed.
- K. Leon, D. Mery, F. Pedreschi and J. Leon, Color measurement in L*a*b* units from RGB digital images, Food Res. Int., 2006, 39, 1084–1091 CrossRef.
- H. Kim, O. Awofeso, S. Choi, Y. Jung and E. Bae, Colorimetric analysis of saliva–alcohol test strips by smartphone-based instruments using machine-learning algorithms, Appl. Opt., 2017, 56, 84–92 CrossRef CAS.
- B. Khanal, P. Pokhrel, B. Khanal and B. Giri, Machine-learning-assisted analysis of colorimetric assays on paper analytical devices, ACS Omega, 2021, 6, 33837–33845 CrossRef CAS PubMed.
- G. M. Fernandes, W. R. Silva, D. N. Barreto, R. S. Lamarca, P. C. F. L. Gomes, J. F. da S Petruci and A. D. Batista, Novel approaches for colorimetric measurements in analytical chemistry–a review, Anal. Chim. Acta, 2020, 1135, 187–203 CrossRef CAS PubMed.
- S.-L. Lee and C.-C. Tseng, Color image enhancement using histogram equalization method without changing hue and saturation, in 2017 IEEE International Conference on Consumer Electronics-Taiwan (ICCE-TW), IEEE, 2017, pp. 305–306 Search PubMed.
- J. A. Sidey-Gibbons and C. J. Sidey-Gibbons, Machine learning in medicine: a practical introduction, BMC Med. Res. Methodol., 2019, 19, 1–18 CrossRef PubMed.
- A. Vaniya and O. Fiehn, Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics, TrAC, Trends Anal. Chem., 2015, 69, 52–61 CrossRef CAS PubMed.
- P. Langley and H. A. Simon, Applications of machine learning and rule induction, Commun. ACM, 1995, 38, 54–64 CrossRef.
- A. Garofalo, C. Di Sarno and V. Formicola, Enhancing intrusion detection in wireless sensor networks through decision trees, in Dependable Computing: 14th European Workshop, EWDC 2013, Coimbra, Portugal, May 15-16, 2013, Springer, 2013, pp. 1–15 Search PubMed.
- M. Rezazadeh, S. Seidi, M. Lid, S. Pedersen-Bjergaard and Y. Yamini, The modern role of smartphones in analytical chemistry, TrAC, Trends Anal. Chem., 2019, 118, 548–555 CrossRef CAS.
- B. T. Kurien and R. H. Scofield, Western blotting, Methods, 2006, 38, 283–293 CrossRef CAS PubMed.
- A. Y. Mutlu, V. Kılıç, G. K. Özdemir, A. Bayram, N. Horzum and M. E. Solmaz, Smartphone-based colorimetric detection via machine learning, Analyst, 2017, 142, 2434–2441 RSC.
- Z. Chen, J. Liu, J. Li, M. Yuan and G. Yu, Leveraging multi-output modelling for CIELAB using colour difference formula towards sustainable textile dyeing, Auton. Intell. Syst., 2024, 4, 1–13 CrossRef.
- M. E. Solmaz, A. Y. Mutlu, G. Alankus, V. Kılıç, A. Bayram and N. Horzum, Quantifying colorimetric tests using a smartphone app based on machine learning classifiers, Sens. Actuators, B, 2018, 255, 1967–1973 CrossRef CAS.
- Y. Xie, H. Zhang, S. Zhang, S. Xiao, Q. Li and X. Qin, A data-driven approach for predicting industrial dyeing recipes of polyester fabrics, Fibers Polym., 2024, 25, 2985–2991 CrossRef CAS.
- C. Ichsan and S. Rodiah, The best performing color space and machine learning regression algorithm for the accurate estimation of chromium(VI) and iron(III) in aqueous samples using low-cost and portable flatbed scanner colorimetry, J. Iran. Chem. Soc., 2024, 21, 2335–2349 CrossRef CAS.
- E. Lopez-Lopez and R. Mendez-Rial, Multilayer perceptron to boost colorimetric capacities of 2D RGB and hyperspectral snapshot cameras, in 2022 10th European Workshop on Visual Information Processing (EUVIP), IEEE, 2022, pp. 1–6 Search PubMed.
- K. Y. Bae, H. S. Jang, B. C. Jung and D. K. Sung, Effect of prediction error of machine learning schemes on photovoltaic power trading based on energy storage systems, Energies, 2019, 12, 1249 CrossRef.
- A. G. Gornall, C. J. Bardawill and M. M. David, Determination of serum proteins by means of the biuret reaction, J. Biol. Chem., 1949, 177, 751–766 CrossRef CAS PubMed.
- N. J. Kruger, The Bradford method for protein quantitation, in The Protein Protocols Handbook, 2009, pp. 17–24 Search PubMed.
- J. M. Walker, The Protein Protocols Handbook, Springer, 2002 Search PubMed.
- G. M. Johnson, X. Song, E. D. Montag and M. D. Fairchild, Derivation of a color space for image color difference measurement, Color Res. Appl., 2010, 35, 387–400 CrossRef.
- M.-Y. Jia, Q.-S. Wu, H. Li, Y. Zhang, Y.-F. Guan and L. Feng, The calibration of cellphone camera-based colorimetric sensor array and its application in the determination of glucose in urine, Biosens. Bioelectron., 2015, 74, 1029–1037 CrossRef CAS PubMed.
- L. Breiman, Random forests, Mach. Learn., 2001, 45, 5–32 CrossRef.
- D. Wang, M. Wang and X. Qiao, Support vector machines regression and modeling of greenhouse environment, Comput. Electron. Agric., 2009, 66, 46–52 CrossRef.
- L. Perotin; A. Défossez; E. Vincent; R. Serizel and A. Guérin, Regression versus classification for neural network based audio source localization, in 2019 IEEE Workshop on Applications of Signal Processing to Audio and Acoustics (WASPAA), IEEE, 2019, pp. 343–347 Search PubMed.
- V. Rodriguez-Galiano, M. Sanchez-Castillo, M. Chica-Olmo and M. Chica-Rivas, Machine learning predictive models for mineral prospectivity: an evaluation of neural networks, random forest, regression trees and support vector machines, Ore Geol. Rev., 2015, 71, 804–818 CrossRef.
- M. Lebendiker and T. Danieli, Production of prone-to-aggregate proteins, FEBS Lett., 2014, 588, 236–246 CrossRef CAS PubMed.
- W. W. Cleland, Dithiothreitol, a new protective reagent for SH groups, Biochemistry, 1964, 3, 480–482 CrossRef CAS PubMed.
- M. P. Deutscher, Maintaining protein stability, Methods Enzymol., 2009, 463, 121–127 CAS.
- P. Perez, C. Hue, J. Vermaak and M. Gangnet, Color-based probabilistic tracking computer vision, in Proceedings 7th European Conferences Computer Vision, Copenhagen, Denmark, 2002, pp. 28–31 Search PubMed.
- W. Xu, K. Fu and P. W. Bohn, Electrochromic sensor for multiplex detection of metabolites enabled by closed bipolar electrode coupling, ACS Sens., 2017, 2, 1020–1026 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Image of manufactured prototype of the color sensor; protein quantitation curve of absorbance at 560 nm vs. protein concentration in H2O of BCA assay and Bradford assay; RGB color component values vs. frequency of color sensor output values; calibration curves for CIELAB, HSL and HSV values; sum of RGB color component values vs. frequency of color sensor outputs, ratio of individual R, G, B colors to the sum of RGB, and ratio of individual C, M, Y, K colors to the sum of CMYK with increasing protein concentration for the Bradford assay test; calibration curves for HSL values; summary of regression model error matrices of various color models; summary of previous color sensor and regression model work reported. See DOI: https://doi.org/10.1039/d4ra07510b |
| This journal is © The Royal Society of Chemistry 2025 |