Open Access Article
Open Access ArticleLigand-free palladium-catalyzed synthesis of 3-(2,2-dialkyl-2H-chromen-4-yl)-2-phenylimidazo[1,2-a]pyridine derivatives: molecular docking investigation of their potential as DNA gyrase inhibitors and evaluation of their antibacterial activities†
Rudra Narayan Mishra a,
Mohammed Ansar Ahemad
a,
Mohammed Ansar Ahemad a,
Jasmine Panda
a,
Jasmine Panda a,
Sabita Nayak
a,
Sabita Nayak *a,
Seetaram Mohapatra
*a,
Seetaram Mohapatra a and
Chita Ranjan Sahoob
a and
Chita Ranjan Sahoob
aOrganic Synthesis Laboratory, Department of Chemistry, Ravenshaw University, Cuttack 753003, Odisha, India. E-mail: sabitanayak18@gmail.com
bICMR-Regional Medical Research Centre, Department of Health Research, Ministry of Health & Family Welfare, Govt. of India, Bhubaneswar 751023, Odisha, India
First published on 30th January 2025
Abstract
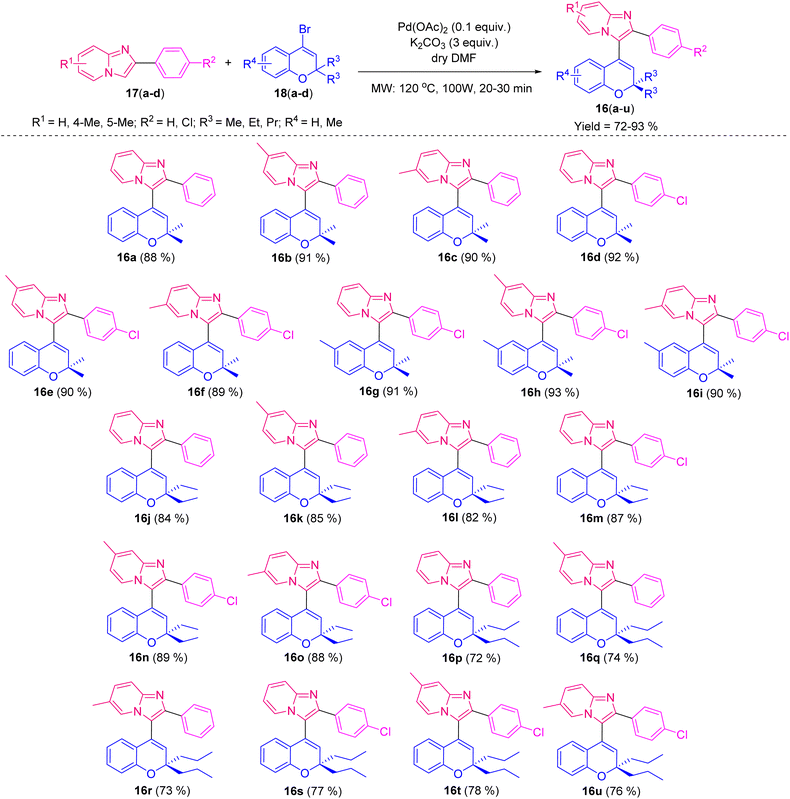
Palladium-catalyzed reactions between imidazo[1,2-a]pyridine derivatives and 4-bromo-2,2-dialkyl-substituted 2H-chromenes under microwave irradiation at 100 W, 120 °C for 20–30 min provided a series of new 3-(2,2-dialkyl-2H-chromen-4-yl)-2-phenylimidazo[1,2-a]pyridine derivatives in good to excellent yields. The structures of the synthesized compounds were confirmed through spectroscopic techniques (NMR and HRMS). The X-ray single-crystal structure of compound 16e was also determined. Shorter reaction time, high yield and good substrate scope were the major advantages of this method. All these compounds were further investigated in vitro for the evaluation of their antibacterial potency using the agar well diffusion method against human pathogenic Gram-negative E. coli and Gram-positive S. aureus bacteria, with the determination of their minimum inhibitory concentration (MIC) values. Indeed, compound 16h strongly inhibited DNA gyrase in silico with a binding affinity of −8.7 kcal mol−1 and exhibited zone of inhibition (ZI) values of 19 mm and MIC values of 10 μg mL−1 in both Gram-negative E. coli and Gram-positive S. aureus, relative to the standard drug gentamicin. By analyzing the structure–activity relationships based on the molecular docking results and the potent antibacterial activities, it could be concluded that these new phenylimidazo[1,2-a]pyridine-chromene derivatives have the potential to be effective druggable antibacterial agents.
1. Introduction
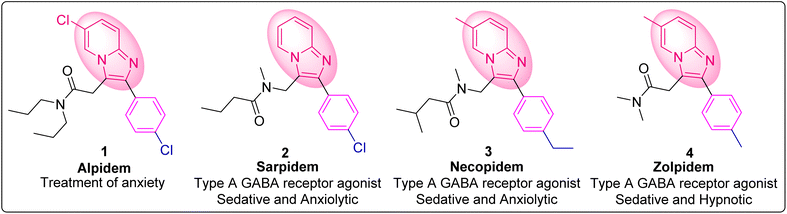
In today's world of global health concerns, antimicrobial resistance (AMR) is a serious, comprehensive and multisectoral issue.1 The global rise in AMR poses a significant threat, as it diminishes the efficacy of common antibiotics against widespread bacterial infections. To combat the issue of antibiotic resistance, a concerted and urgent effort is required to develop new antibiotics with improved toxicity profiles. In this regard, heterocyclic moieties are very important in medicinal chemistry.2 Hence, this quest has provoked scientists to investigate a wide range of physiologically active heterocycles in order to create effective antibacterial drug candidates.Among numerous heterocycles, imidazo[1,2-a]pyridine (IZP) is the backbone of various marketed drugs, such as alpidem, saripidem, necopidem, zolpidem, olprinone, and zolimidine (Fig. 1).3–5 IZP is a multitargeted scaffold possessing a wide range of pharmacological activities, such as antibacterial, anticancer, anti-inflammatory, antihypertensive, antiviral, antiosteoporotic, antiparasitic, and neuroprotective properties.6–10 IZPs can target several enzymes associated with the synthesis of cell wall/peptidoglycan, protein, folic acid, DNA or RNA to eradicate infections. Investigations to date have shown that substituted IZP rings exhibit remarkable antibacterial activity.11
IZPs bearing bis-heterocycles show good antibacterial activity, as reported in various literature sources.11 For instance, Al-Tel et al. (2011) reported the synthesis of indole-based-IZPs 5 from 6 and 7 (see Fig. 2a) as antibacterial agents. These compounds exhibited strong inhibition against S. aureus, E. faecalis, B. megaterium, E. coli, P. aeruginosa and E. aerogenes, compared to control antibiotics, such as cefixime and amoxicillin. The nature of the substituents, such as aryl groups, exemplified the extent of the activity of these compounds (Fig. 2a).12,13 In 2019, Ebenezer et al. reported the synthesis of pyrazole-IZP molecular conjugates 8 as antibacterial agents targeting cell wall synthesis from aminopyridine 9, alkyne 10 and pyrazole aldehyde 11 using CuSO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sodium ascorbate
sodium ascorbate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cs2CO3 in a ratio of 1
Cs2CO3 in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 at 120 °C for 12 h.14 The compounds displayed pronounced bactericidal activity, as demonstrated by their MIC and time-kill kinetics values (Fig. 2b).15 Thakur et al. (2020) reported the synthesis of IZP-bearing pyran bis-heterocyclic derivatives 12 as potent antibacterial agents from phenylimidazo[1,2-a]pyridine-3-carbaldehyde 13, 5,5-dimethylcyclohexane-1,3-dione 14, and malononitrile 15 in 5 mL GAAS
0.2 at 120 °C for 12 h.14 The compounds displayed pronounced bactericidal activity, as demonstrated by their MIC and time-kill kinetics values (Fig. 2b).15 Thakur et al. (2020) reported the synthesis of IZP-bearing pyran bis-heterocyclic derivatives 12 as potent antibacterial agents from phenylimidazo[1,2-a]pyridine-3-carbaldehyde 13, 5,5-dimethylcyclohexane-1,3-dione 14, and malononitrile 15 in 5 mL GAAS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL EtOH at 80 °C. Indeed, antibacterial studies showed significant results for the synthesized compounds against E. coli, S. aureus, and S. typhi (Fig. 2c).16
1 mL EtOH at 80 °C. Indeed, antibacterial studies showed significant results for the synthesized compounds against E. coli, S. aureus, and S. typhi (Fig. 2c).16
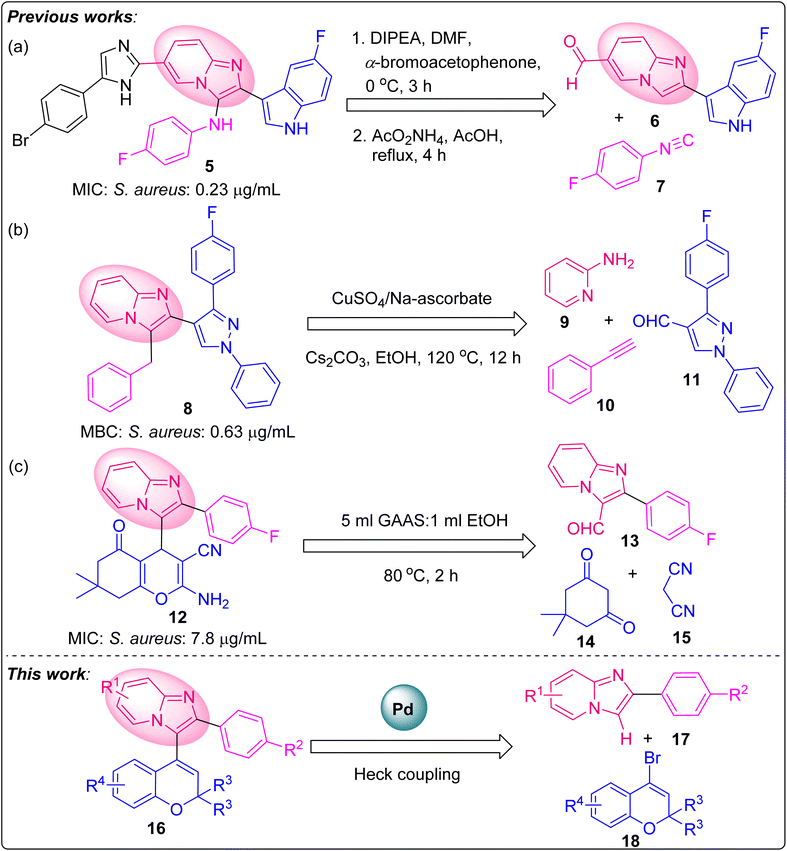 | ||
| Fig. 2 Imidazo[1,2-a]pyridine-based molecules exhibiting potent antibacterial activity against Gram-positive S. aureus. | ||
In the quest to develop new antibacterial compounds, our group in 2021 reported 2H-chromene-based IZP derivatives as potent peptide deformylase inhibitors from chromene aldehyde and 2-aminopyridine in FeCl3, and MeNO2/DMF under microwave irradiation at 60 W, 100 °C in 15 min. The synthesized compounds exhibited potent antibacterial activity against bacterial strains such as K. oxytoca, S. pyogenes, S. aureus, and E. coli.17 They were synthesized as monosubstituted imidazo[1,2-a]pyridine, but we had tried to synthesize disubstituted imidazo[1,2-a]pyridine. In our attempts, we tried various different substituents in the phenyl ring of imidazo[1,2-a]pyridine moiety and also in the chromene core.
Keeping this idea in mind, in this study we planned to fuse both 4-bromo-2,2-dialkyl-substituted 2H-chromenes and the imidazo[1,2-a]pyridine moiety in a single molecular entity to construct phenylimidazo[1,2-a]pyridine-based chromene hybrid molecules via a palladium-catalyzed cross-coupling reaction. To the best of our knowledge, this is the first report on the synthesis and antibacterial activity of 3-(2,2-dialkyl-2H-chromen-4-yl)-2-phenylimidazo[1,2-a]pyridine derivatives. Molecular docking investigations were further employed to gain better insights into the hybrid's potency while attempting to provide a mechanism for the action of the hybrid compounds as DNA gyrase inhibitors.
2. Results and discussion
2.1 Chemistry
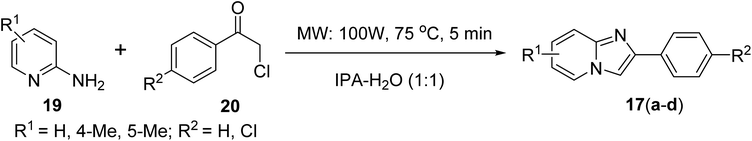
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) under microwave heating at 100 W at 75 °C for 5 min (Scheme 1) following a known literature procedure.18,19
1) under microwave heating at 100 W at 75 °C for 5 min (Scheme 1) following a known literature procedure.18,19
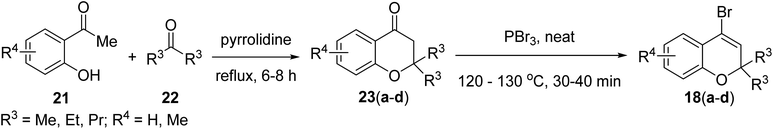
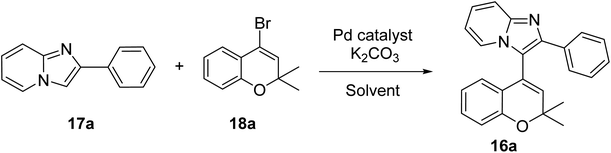
After having 17(a–d) in hand, we further synthesized compounds 18(a–d). Initially, o-hydroxy acetophenone 21 and substituted ketones 22 reacted by aldol condensation followed by intramolecular cyclization to provide substituted 2H-chromones 23, which then underwent reaction with PBr3 to give 4-bromo-2,2-dialkyl-substituted 2H-chromenes 18(a–d) (Scheme 2).20,21
After the successful synthesis of the starting materials 17a and 18a, we further proceeded towards the synthesis of 16a. Very recently, Raiguru et al. from our group18 reported in 2023 the synthesis of imidazo[1,2-a]pyridine-flavone hybrid derivatives using imidazo[1,2-a]pyridine and bromo flavone by a Heck coupling reaction using Pd(OAc)2 in 1,4-dioxane and ethanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 100 W at 100 °C for 30 min using TPP as a ligand. We applied the same conditions for the coupling of 17a and 18a, but unfortunately failed to achieve the coupled product. In order to achieve 16a by the coupling reaction between 17a and 18a, we screened the reaction conditions by trying various palladium catalysts, bases, ligands, and solvents.
1) at 100 W at 100 °C for 30 min using TPP as a ligand. We applied the same conditions for the coupling of 17a and 18a, but unfortunately failed to achieve the coupled product. In order to achieve 16a by the coupling reaction between 17a and 18a, we screened the reaction conditions by trying various palladium catalysts, bases, ligands, and solvents.
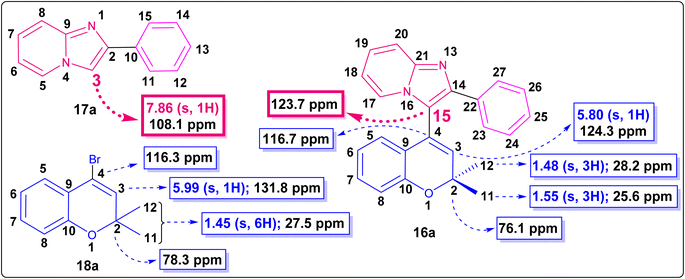
The reaction was carried out under both conventional heating as well as microwave irradiation. From Table 1, it can be seen that the optimal reaction conditions were Pd(OAc)2, K2CO3 in DMF at 100 W at 120 °C for 20 min, which provided the product 16a in an 88% yield (Table 1, entry 10). The results obtained from both the conventional and microwave irradiation methods are summarized in Table 1. The product 16a was characterized by 1H, 13C NMR, and HRMS analyses.
| Entrya | Coupling reagent | Base | Solvent | Ligand | Conventional method | Microwave irradiation method | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Temp. (°C) | Time (h) | Yieldb (%) | MW (W) | Temp (°C) | Time (min) | Yieldb (%) | |||||
| a Reaction conditions: 17a (1 mmol), 18a (3 mmol), Pd catalyst (0.1 equiv.), K2CO3 as base (3 equiv.), and solvent (3–4 mL).b Isolated yield, MW: microwave. | |||||||||||
| 1 | Pd(OAc)2 | — | 1,4-Dioxane/EtOH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
TPP | 100 | 3 | — | 100 | 100 | 30 | — |
| 2 | Pd(PPh)3Cl2 | Et3N | DMF | — | 100 | 4 | 11 | 50 | 80 | 30 | 44 |
| 3 | Pd(PPh)3Cl2 | K3PO4 | DMF | — | 100 | 4 | 24 | 70 | 90 | 30 | 36 |
| 4 | Pd(PPh)3Cl2 | K2CO3 | DMF | — | 100 | 4 | 27 | 90 | 100 | 30 | 30 |
| 5 | Pd(PPh)3Cl2 | NaOAc | DMF | — | 100 | 4 | 10 | 120 | 120 | 30 | 32 |
| 6 | Pd(OAc)2 | Et3N | DMF | TPP | 120 | 4 | 24 | 80 | 100 | 20 | 40 |
| 7 | Pd(OAc)2 | K3PO4 | DMF | — | 100 | 3 | 31 | 100 | 120 | 20 | 40 |
| 8 | Pd(OAc)2 | K2CO3 | DMF | — | 120 | 2 | 76 | 80 | 100 | 10 | 75 |
| 9 | Pd(OAc)2 | NaOAc | DMF | — | 100 | 4 | 23 | 90 | 100 | 20 | 65 |
| 10 | Pd(OAc)2 | K2CO3 | DMF | — | 150 | 5 | 54 | 100 | 120 | 20 | 88 |
| 11 | Pd(OAc)2 | K2CO3 | CH3CN | — | 80 | 4 | 36 | 100 | 120 | 20 | 70 |
After optimizing the reaction conditions and characterization of 16a by 1H, 13C NMR, and HRMS, we wished to expand the substrate scope of the phenylimidazo[1,2-a]pyridine-chromene derivatives. Consequently, the substituted imidazo[1,2-a]pyridines 17(a–d) were coupled with four different 4-bromo-2,2-dialkyl substituted 2H-chromene derivatives 18(a–d), furnishing 21 phenylimidazo[1,2-a]pyridine-chromene derivatives 16(a–u) with good yields, as shown in Table 2.
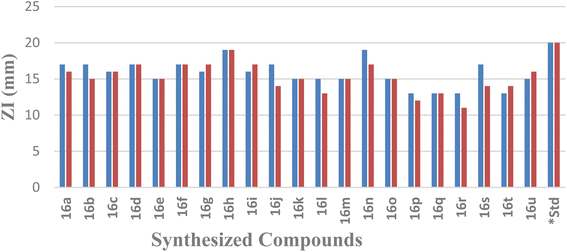
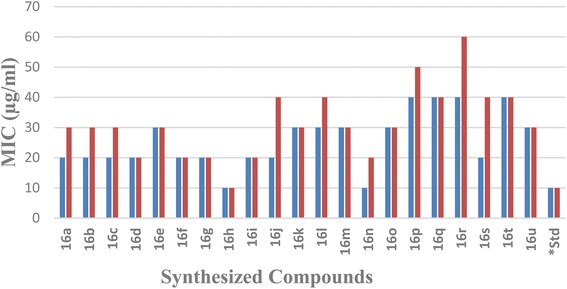
| Compound code | E. coli | S. aureus | ||
|---|---|---|---|---|
| ZI (mm) | MIC (μg ml−1) | ZI (mm) | MIC (μg ml−1) | |
| a ZI: zone of inhibition, MIC: minimum inhibitory concentration, *standard drug: gentamicin. | ||||
| 16a | 17 | 20 | 16 | 30 |
| 16b | 17 | 20 | 15 | 30 |
| 16c | 16 | 20 | 16 | 30 |
| 16d | 17 | 20 | 17 | 20 |
| 16e | 15 | 30 | 15 | 30 |
| 16f | 17 | 20 | 17 | 20 |
| 16g | 16 | 20 | 17 | 20 |
| 16h | 19 | 10 | 19 | 10 |
| 16i | 16 | 20 | 17 | 20 |
| 16j | 17 | 20 | 14 | 40 |
| 16k | 15 | 30 | 15 | 30 |
| 16l | 15 | 30 | 13 | 40 |
| 16m | 15 | 30 | 15 | 30 |
| 16n | 19 | 10 | 17 | 20 |
| 16o | 15 | 30 | 15 | 30 |
| 16p | 13 | 40 | 12 | 50 |
| 16q | 13 | 40 | 13 | 40 |
| 16r | 13 | 40 | 11 | 60 |
| 16s | 17 | 20 | 14 | 40 |
| 16t | 13 | 40 | 14 | 40 |
| 16u | 15 | 30 | 16 | 30 |
| *Standard | 20 | 10 | 20 | 10 |
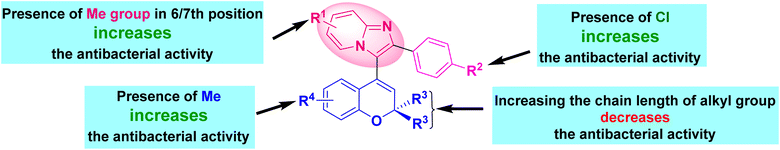
It was found that the yield of the product depended on the various substituents at the different positions. In the chromene core, whenever the alkyl chain length at the 2-position increased, the yield gradually decreased. Trisubstituted methyl groups in the chromene ring 16(g–i) also provided better yields than for the 2,2-dimethyl-substituted chromene. In the case of imidazo[1,2-a]pyridine, whenever there was chloro substitution in the benzene ring, i.e., 16(d–i), 16(m–o), and 16(s–u), the yields were better than the unsubstituted ones, i.e., 16a, 16j, and 16p. Whenever the methyl substituents was in the 7th position of imidazo[1,2-a]pyridine (16b, 16e, 16h, 16k, 16n, 16q, and 16t), the yield was better than the 6th-positioned methyl imidazo[1,2-a]pyridine (16c, 16f, 16i, 16l, 16o, 16r, and 16u). We also tried the reaction with keeping the methyl group in the 5th position of imidazo[1,2-a]pyridine, but a series of spots developed, preventing us from separating out the product.
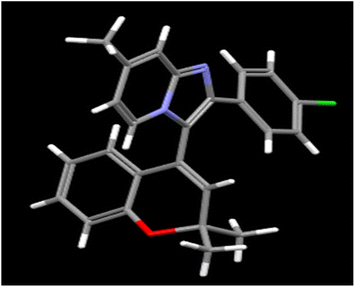 | ||
| Fig. 4 Single-crystal X-ray diagram of 2-(4-chlorophenyl)-3-(2,2-dimethyl-2H-chromen-4-yl)-7-methylimidazo[1,2-a]pyridine 16e (CCDC 2347891). | ||
2.2 Biological studies
| Compound code | E. coli DNA gyrase (PDB ID: 1AJ6) | S. aureus DNA gyrase (PDB ID: 3G7B) | ||
|---|---|---|---|---|
| Binding affinity (kcal mol−1) | Residues showing interaction | Binding affinity (kcal mol−1) | Residues showing interaction | |
| 16a | −8.7 | ASP 34, ASP 38, GLU 31, ASN 35, ILE 67, PRO 68, VAL 97, ILE 78, SER 98, ALA 84, VAL 81, GLY 96, HIS 83, LEU 82, LYS 87, PHE 88, GLY 86, GLY 94 | −8.1 | ILE 129, ASN 31, ILE 79, THR 127, SER 32, ASP 58, GLY 62, ILE 63, GLU 35, PRO 64, ARG 98, ARG 61, ASP 34 |
| 16b | −8.6 | ALA 42, ARG 65, ASP 38, GLU 39, ILE 67, ALA 36, ASP 62, THR 142, ASP 34, ASN 35, GLY 94, GLU 31, GLY 96, VAL 97, ILE 78 | −7.5 | ASN 31, ASP 34, GLY 62, ARG 61, GLU 35, ARG 98, PRO 64, ILE 63, THR 127, SER 32, ASP 58, ILE 129, ILE 79, LEU 80, SER 83 |
| 16c | −8.5 | ALA 42, ASP 38, ARG 65, GLU 39, ILE 67, ALA 36, ASP 62, THR 142, ASP 34, ASN 35, MET 79, GLY 94, GLU 31, VAL 97, GLY 96, ILE 78 | −7.6 | ILE 79, ASN 31, SER 32, ASP 58, THR 127, ILE 63, GLY 62, GLU 35, PRO 64, ARG 98, ARG 61, ALA 38, ASP 34 |
| 16d | −8.4 | ILE 67, ASP 62, THR 142, ASN 35, VAL 97, ILE 78, GLY 96, HIS 83, ALA 84, ASP 38, ALA 42, GLU 39, ARG 65, PRO 68 | −8.1 | ILE 28, ILE 79, ILE 129, THR 127, ASN 31, SER 32, ASP 58, ILE 63, GLY 62, GLU 35, PRO 64, ARG 98, ARG 61, ASP 34 |
| 16e | −7.2 | PRO 68, ILE 67, ARG 65, MET 79, ASN 35, THR 142, VAL 144, VAL 97, VAL 32, ASP 62, ALA 36, ILE 78, GLU 39, GLY 96, ASP 34, ASP 38 | −7.6 | ILE 79, ARG 98, ARG 61, PRO 64, GLY 62, GLU 35, ILE 63, THR 127, SER 32, ASP 58, ASN 31, ALA 38, ASP 34 |
| 16f | −8.4 | ILE 67, GLU 39, ARG 65, ASP 38, ALA 42, ASP 34, GLY 94, GLU 31, ASN 35, GLY 96, VAL 97, ILE 78, THR 142, MET 79 | −8.0 | PRO 64, GLU 35, ILE 63, ARG 61, GLY 62, THR 127, ASP 58, SER 32, ILE 129, ILE 28, ASN 31, LEU 80, SER 83, ILE 79, ASP 34 |
| 16g | −8.1 | ALA 42, ASP 38, ALA 84, HIS 83, GLY 96, SER 98, VAL 97, ILE 78, GLU 39, ASN 35, THR 142, ASP 62, ILE 67, ALA 36, PRO 68, ARG 65 | −8.4 | ASP 34, ALA 38, GLU 35, ARG 98, PRO 64, GLY 62, ARG 61, ASP 58, ILE 63, THR 127, SER 32, ILE 79, ASN 31 |
| 16h | −8.7 | ASP 34, ASP 38, VAL 144, MET 79, VAL 32, THR 142, ALA 36, ASP 62, GLU 39, ASN 35, ILE 67, VAL 97, ALA 74, PRO 68, ILE 78, VAL 81, SER 98, HIS 83, ALA 84, GLY 96, GLY 94 | −8.7 | ARG 61, ARG 98, PRO 64, GLY 62, GLU 35, ILE 63, ASP 58, THR 127, SER 32, ASN 31, ASP 34, ALA 38, ILE A:79 |
| 16i | −8.4 | ASP 38, ARG 65, ALA 42, HIS 83, ALA 84, GLY 96, VAL 97, SER 98, ILE 78, GLU 39, ASN 35, THR 142, ILE 67, ASP 62, ALA 36, GLY 66, ALA 75, GLY 64, PRO 68, ALA 74 | −8.5 | ILE 79, ASN 31, ILE 129, SER 32, ASP 58, THR 127, GLY 62, ILE 63, GLU 35, PRO 64, ARG 98, ARG 61, ALA 38, ASP 34 |
| 16j | −8.5 | VAL 97, SER 98, GLY 96, ILE 78, ARG 65, PRO 68, GLU 39, GLY 66, ILE 67, ASP 62, THR 142, ALA 36, ASN 35, VAL 32, VAL 144, MET 79, ASP 38 | −7.1 | ILE 79, PRO 64, ARG 98, ARG 61, GLY 62, GLU 35, ILE 63, THR 127, ASP 58, ASN 31, ALA 38, ASP 34 |
| 16k | −7.7 | PRO 68, ALA 84, HIS 83, ILE 78, GLY 96, ASN 35, VAL 97, ILE 67, MET 79, VAL 144, THR 142, ASP 62, ALA 36, ASP 38, ALA 42, GLU 39, GLY 66, ARG 65 | −7.7 | PRO 64, ARG 98, ARG 61, GLY 62, GLU 35, ILE 63, ASP 58, SER 32, THR 127, ILE 129, ASP 34, ALA 38, ASN 31, ILE 79 |
| 16l | −8.0 | ILE 78, PRO 68, ILE 67, GLU 39, ALA 36, ASP 62, THR 142, ASP 38, ASN 35, VAL 144, MET 79, HIS 83, ALA 84, VAL 97, VAL 81, SER 98, GLY 96 | −7.5 | ILE 79, THR 127, ILE 63, GLU 35, ASP 34, ASN 31, ASP 30, GLU 27, VAL 84, ILE 129, SER 83, SER 82, LEU 80, ILE 28 |
| 16m | −8.0 | PRO 68, GLY 66, ALA 36, ASP 62, ILE 67, THR 142, VAL 144, MET 79, ASN 35, VAL 97, ILE 78, GLY 96, ALA 84, ASP 38, ARG 65, GLU 39, ALA 42 | −7.5 | ILE 79, ARG 61, ARG 98, PRO 64, GLU 35, ASN 31, ILE 63, THR 127, GLY 62, ILE 129, SER 32, ASP 34, ASP 58 |
| 16n | −8.7 | ASP 34, ASP 38, GLU 31, ASN 35, ILE 67, ILE 78, MET 79, VAL 97, PRO 68, SER 98, VAL 81, ALA 84, GLY 96, HIS 83, LEU 82, GLY 86, LYS 87, PHE 88, GLY 94 | −7.8 | ILE 79, ASP 34, ILE 63, SER 32, GLU 35, ASN 31, THR 127, GLU 27, ILE 129, ILE 28, LEU 80, VAL 85, VAL 84, SER 83, SER 82 |
| 16o | −8.1 | PRO 68, ARG 65, GLU 39, GLY 66, ILE 67, ASP 62, ALA 36, THR 142, ALA 42, VAL 32, ASP 38, ASN 35, VAL 97, ILE 78 | −7.8 | ARG 98, GLU 35, PRO 64, GLY 62, ARG 61, THR 127, ASP 58, ASN 31, SER 32, ILE 63, ILE 79, ASP 34 |
| 16p | −8.0 | ALA 42, ASP 38, ARG 65, ILE 67, GLU 39, ASP 62, THR 142, ASP 34, ASN 35, GLU 31, GLY 94, VAL 97, GLY 96, SER 98, ALA 84, ILE 78, HIS 83 | −6.9 | ALA 38, ASP 34, ARG 61, ARG 98, PRO 64, GLU 35, GLY 62, ILE 63, ILE A:79, GLY 60, ASN 31, SER 32, ASP 58, THR 127 |
| 16q | −7.2 | ALA 42, ASP 38, ARG 65, GLU 39, GLY 66, ASP 62, THR 142, ILE 67, ASN 35, ILE 78, VAL 97, GLY 96, PRO 68, ALA 74 | −7.2 | ARG 98, ARG 61, GLU 35, ILE 63, PRO 64, GLY 62, GLY 126, ASN 31, GLY 60, ASP 58, THR 127, SER 32, ASP 34, ALA 38, ILE 79 |
| 16r | −7.3 | GLY 66, PRO 68, ALA 84, ILE 78, ILE 67, THR 142, ASN 35, VAL 97, GLY 96, MET 79, VAL 144, ASP 62, ALA 36, ASP 38, ALA 42, ARG 65, GLU 39 | −6.6 | ILE 129, THR 127, ASP 58, ALA 38, SER 32, ASP 34, ILE 79, ARG 61, ARG 98, PRO 64, GLU 35, ASN A:31, GLY 62, ILE 63 |
| 16s | −8.1 | PRO 68, GLY 66, ILE 67, ALA 36, ASP 62, THR 142, ASN 35, VAL 97, GLY 96, HIS 83, ALA 84, ILE 78, ASP 38, ALA 42, ALA 74 | −6.9 | ILE 79, ARG 61, PRO 64, ARG 98, GLU 35, GLY 62, ILE 63, SER 32, THR 127, ASP 58, ASP 34, ALA 38, ASN 31 |
| 16t | −7.7 | ILE 78, VAL 97, VAL 81, HIS 83, ALA 84, GLY 96, ASN 35, GLU 31, GLY 94, ARG 65, ASP 34, PRO 68, GLY 66, THR 142, ILE 67, GLU 39, ALA 36, ASP 38, ALA 42 | −7.4 | ASP 34, ALA 38, ILE 79, ASN 31, ILE 63, SER 32, GLU 35, GLY 62, THR 127, ASP 58, GLY 126, GLY 60, PRO 64, ARG 61, LEU 37 |
| 16u | −7.1 | GLY 66, GLY 64, PRO 68, ASP 62, THR 142, ILE 67, ASN 35, ILE 78, ALA 84, ASP 38, ALA 42, ARG 65, GLU 39 | −7.5 | PRO 64, ARG 61, ARG 98, GLU 35, GLY 62, THR 127, ASP 58, SER 32, ASP 34, ASN 31, ILE 63, ILE 79 |
Compound 16h, which was found to be the most potent, demonstrated a range of ligand–protein interactions, such as van der Waals interactions with the amino acid residues ASP 34, ASP 38, MET 79, VAL 32, THR 142, ALA 36, ASP 62, GLU 39, ASN 35, VAL 97, ALA 74, VAL 81, SER 98, HIS 83, GLY 96, and GLY 94, alkyl and π-alkyl interactions with VAL 144, PRO 68, ALA 84, and π–σ interactions with ILE 67 and ILE 78. According to these findings, these results show the effective insertion of 16h into the active cavities of E. coli DNA gyrase. Similarly, compound 16h also demonstrated a strong binding affinity with an energy of −8.7 kcal mol−1, and was retained within the active pockets of S. aureus DNA gyrase enzymes through van der Waals interactions with ARG 98, GLY 62, ASP 58, THR 127, SER 32, ASN 31, ASP 34, π-cation and π-anion interactions with ARG 61, GLU 35, and alkyl and π-alkyl interactions with ILE 79, PRO 64, ILE 63, and ALA 38. According to the 3D-interaction analysis of the synthesized phenylimidazo[1,2-a]pyridine-chromene analogues, compound 16h containing –Cl in the phenyl ring of the imidazo[1,2-a]pyridine moiety along with trisubstituted methyl in the chromene ring exhibited the highest level of DNA gyrase-inhibitory activity. It also displayed strong antibacterial activity. Fig. 8 and 9 display the corresponding docking images and 3D interactions of compound 16h against DNA gyrases from E. coli (PDB ID: 1AJ6) and S. aureus (PDB ID: 3G7B), respectively.
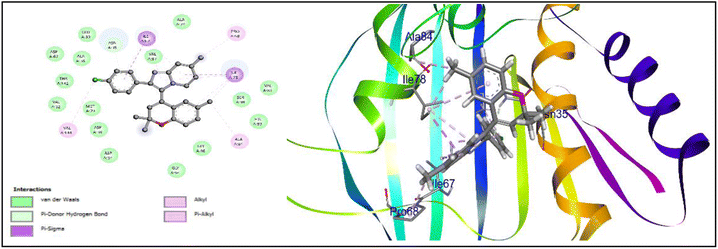 | ||
| Fig. 8 Docking interaction and 3D binding image of compound 16h with E. coli DNA gyrase (PDB ID: 1AJ6). | ||
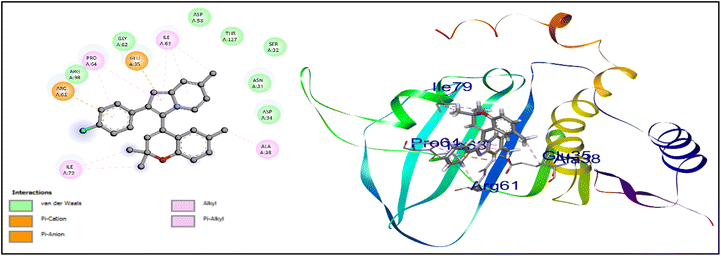 | ||
| Fig. 9 Docking interaction and 3D binding image of compound 16h with S. aureus DNA gyrase (PDB ID: 3G7B). | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 4.15, and BS = 0.55. Additional factors like ROTB < 9 and TPSA < 140 Å2 also influence the bioavailability. The topological polar surface area (TPSA), which is inversely related to the percentage absorption (%ABS = 109 − 0.345 × TPSA), can be used as an alternative to counting the hydrogen-bonding groups for estimating absorption. Here, compound 16a showed zero violations, while gentamicin violated two. Most compounds in the series violated at least one rule, making them suitable drug-like candidates. Five compounds exhibited high gastrointestinal absorption, suggesting effective absorption, with all the compounds achieving systemic circulation (bioavailability = 0.55).
P < 4.15, and BS = 0.55. Additional factors like ROTB < 9 and TPSA < 140 Å2 also influence the bioavailability. The topological polar surface area (TPSA), which is inversely related to the percentage absorption (%ABS = 109 − 0.345 × TPSA), can be used as an alternative to counting the hydrogen-bonding groups for estimating absorption. Here, compound 16a showed zero violations, while gentamicin violated two. Most compounds in the series violated at least one rule, making them suitable drug-like candidates. Five compounds exhibited high gastrointestinal absorption, suggesting effective absorption, with all the compounds achieving systemic circulation (bioavailability = 0.55).
| MWa | M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pb Pb |
ROTBc | HBAd | HBDe | TPSAf | Violations | GI absg | BSh | |
|---|---|---|---|---|---|---|---|---|---|
| a Molecular weight in g mol−1.b Partition coefficient (lipophilicity).c Number of rotatable bonds.d Number of H-bond acceptors.e Number of H-bond donors.f Topological polar surface area in Å2.g Gastrointestinal absorption.h Bioavailability score, *std: gentamicin as the standard. | |||||||||
| 16a | 352.43 | 3.95 | 2 | 2 | 0 | 26.53 | 0 | High | 0.55 |
| 16b | 366.45 | 4.15 | 2 | 2 | 0 | 26.53 | 1 | High | 0.55 |
| 16c | 366.45 | 4.15 | 2 | 2 | 0 | 26.53 | 1 | High | 0.55 |
| 16d | 386.87 | 4.42 | 2 | 2 | 0 | 26.53 | 1 | High | 0.55 |
| 16e | 400.9 | 4.63 | 2 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16f | 400.9 | 4.63 | 2 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16g | 400.9 | 4.63 | 2 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16h | 414.93 | 4.83 | 2 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16i | 414.93 | 4.83 | 2 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16j | 380.48 | 4.36 | 4 | 2 | 0 | 26.53 | 1 | High | 0.55 |
| 16k | 394.51 | 4.56 | 4 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16l | 394.51 | 4.56 | 4 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16m | 414.93 | 4.83 | 4 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16n | 428.95 | 5.03 | 4 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16o | 428.95 | 5.03 | 4 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16p | 408.53 | 4.76 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16q | 422.56 | 4.95 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16r | 422.56 | 4.95 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16s | 442.98 | 5.22 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16t | 457.01 | 5.41 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| 16u | 457.01 | 5.41 | 6 | 2 | 0 | 26.53 | 1 | Low | 0.55 |
| *Std | 477.6 | −2.92 | 7 | 12 | 8 | 199.73 | 2 | Low | 0.17 |
Furthermore, the physicochemical properties and medicinal chemistry of the most potent antibacterial compound, 16h, were assessed using ADMETlab 3.0 to evaluate its ADMET (absorption, distribution, metabolism, excretion, and toxicity) profile, which can help in determining its drug-likeness (Table 5).24,25
| Compound 16h | Recommended limit | |
|---|---|---|
| a Abbreviations: nHA: number of hydrogen-bond acceptors, nHD: number of hydrogen-bond donors, TPSA: topological polar surface area (in Å2), nRig: number of rigid bonds, nRing: number of rings, MaxRing: number of atoms in the biggest ring, %ABS: percentage absorption. | ||
| Physicochemical property | ||
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
6.475 | 0 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log octanol/water log octanol/water |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
−7.094 | −4 to 0.5 mol l−1 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D D |
5.292 | 1 to 3 |
| nHA | 3.0 | 0 to 12 |
| nHD | 0.0 | 0 to 7 |
| TPSA | 26.53 | 0 to 140 Å2 |
| nRot | 2.0 | 0 to 11 |
| nRing | 5.0 | 0 to 6 |
| MaxRing | 10.0 | 0 to 18 |
| nRig | 27.0 | 0 to 30 |
| nHet | 4.0 | 1 to 15 |
| fChar | 0.0 | −4 to 4 |
| %ABS | 99.84% | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Medicinal chemistry | ||
| MCE-18 | 59.871 | MCE-18 ≥ 45 |
| NP score | −0.45 | −5 to 5 |
| Lipinski rule | Accepted | MW ≤ 500, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 5, nHA ≤ 10, nHD ≤ 5, TPSA ≤ 140 Å2 P ≤ 5, nHA ≤ 10, nHD ≤ 5, TPSA ≤ 140 Å2 |
| Pfizer rule | Rejected | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 3, TPSA ≥ 75 Å2 P ≤ 3, TPSA ≥ 75 Å2 |
| Golden triangle | Rejected | 200 ≤ MW ≤ 500, −2 ≤ log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D ≤ 5 D ≤ 5 |
The ADMET characteristics of the most potent compound 16h are presented in Table 6. The compound demonstrated favourable results in terms of human intestinal absorption (HIA) and the predicted bioavailability (F50%) score. Regarding the metabolism, the Caco-2 permeability value was −4.578![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log units. The prediction indicated a relatively low metabolic clearance (CL = 4.967 mL min−1 kg−1) with a short half-life (T1/2 = 0.541 h). Therefore, based on the ADMET analysis, compound 16h is expected to be safe and is unlikely to cause eye corrosion or irritation.
log units. The prediction indicated a relatively low metabolic clearance (CL = 4.967 mL min−1 kg−1) with a short half-life (T1/2 = 0.541 h). Therefore, based on the ADMET analysis, compound 16h is expected to be safe and is unlikely to cause eye corrosion or irritation.
| ADMET | Compound 16h | Recommended limit |
|---|---|---|
| a MDCK: apparent permeability in Madin–Darby canine kidney cells, HIA: human intestinal absorption, VD: volume of distribution, F50%: 50% bioavailability, PPB: plasma protein binding, BBB: blood–brain barrier, PGP: plasma glycoprotein, CL: clearance, DILI: drug-induced liver injury, T1/2: half-life period. | ||
| Absorption | ||
| Caco-2 permeability | −4.578 | ≥−5.15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log unit log unit |
| MDCK permeability | −4.56 | ≤20 × 10−6 cm s−1 |
| Pgp-inhibitor | 0.998 | 0 to 0.5 |
| Pgp-substrate | 0.008 | 0 to 0.5 |
| HIA | 0.0 | |
| F50% | 0.004 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Distribution | ||
| PPB | 99.266% | ≤90% |
| BBB | 0.999 | <0.5 |
| VD | 0.377 | 0.04 to 20 l kg−1 |
| Fu | 0.409% | 5% to 20% |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Metabolism | ||
| CYP1A2 inhibitor | 1.0 | 0 to 0.5 |
| CYP1A2 substrate | 0.997 | 0 to 0.5 |
| CYP2C19 inhibitor | 1.0 | 0 to 0.5 |
| CYP2C19 substrate | 0.986 | 0 to 0.5 |
| CYP2C9 inhibitor | 1.0 | 0 to 0.5 |
| CYP2C9 substrate | 0.922 | 0 to 0.5 |
| CYP2D6 inhibitor | 0.03 | 0 to 0.5 |
| CYP2D6 substrate | 0.047 | 0 to 0.5 |
| CYP3A4 inhibitor | 0.723 | 0 to 0.5 |
| CYP3A4 substrate | 0.996 | 0 to 0.5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Excretion | ||
| CL | 4.967 | 5 to 15 mL min−1 kg−1 |
| T1/2 | 0.541 | >4 h |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Toxicity | ||
| DILI | 0.907 | <0.5 |
| Skin sensitization | 0.682 | <0.5 |
| Eye corrosion | 0.0 | <1 |
| Eye irritation | 0.277 | <1 |
3. Conclusion
In conclusion, a series of 21 novel 3-(2,2-dialkyl-2H-chromen-4-yl)-2-phenylimidazo[1,2-a]pyridine derivatives were produced in good to excellent yields by a proficient palladium-catalyzed cross-coupling reaction under microwave irradiation. The antibacterial activities of all these compounds were assessed in vitro using the agar well diffusion method against human pathogenic Gram-negative E. coli and Gram-positive S. aureus bacteria, and their MIC values were determined. Compound 16h displayed ZI values of 19 mm and MIC values of 10 μg mL−1 in both E. coli and S. aureus, in comparison to the standard drug gentamicin. It also strongly inhibited DNA gyrase in silico with a binding affinity of −8.7 kcal mol−1. As inferred from the structure–activity relationships based on molecular docking studies and their strong antibacterial properties, these novel phenylimidazo[1,2-a]pyridine-chromene derivatives may be powerful antibacterial agents for use in the near future. Therefore, we anticipate that our work will aid in the creation of novel heterocyclic scaffolds by the synthetic fraternity, with a focus on producing effective antibacterial candidates for drug development.4. Experimental section
4.1 Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The reaction mixture was irradiated under microwave heating at 100 W for 5 min at 75 °C. The progress of the reaction was monitored by TLC. After completion of the reaction, the reaction mixture was ice cooled to give a solidified product. The filtered solid product was then recrystallised from IPA to afford the pure desired 2,4′-dichloro-imidazo[1,2-a]pyridine derivatives. However, in the case of 2-chloroacetophenone, the reaction mixture was extracted with ethyl acetate (10 mL, twice), then the combined organic layer was treated over anhydrous Na2SO4. The combined filtrate was subjected to evaporation to obtain the crude compound, which was purified by silica gel column chromatography (60–120 mesh) using 30% ethyl acetate in hexane as the eluent to obtain the corresponding phenyl-substituted imidazo[1,2-a]pyridine as product 17 (Scheme 1).18,19
1). The reaction mixture was irradiated under microwave heating at 100 W for 5 min at 75 °C. The progress of the reaction was monitored by TLC. After completion of the reaction, the reaction mixture was ice cooled to give a solidified product. The filtered solid product was then recrystallised from IPA to afford the pure desired 2,4′-dichloro-imidazo[1,2-a]pyridine derivatives. However, in the case of 2-chloroacetophenone, the reaction mixture was extracted with ethyl acetate (10 mL, twice), then the combined organic layer was treated over anhydrous Na2SO4. The combined filtrate was subjected to evaporation to obtain the crude compound, which was purified by silica gel column chromatography (60–120 mesh) using 30% ethyl acetate in hexane as the eluent to obtain the corresponding phenyl-substituted imidazo[1,2-a]pyridine as product 17 (Scheme 1).18,19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) as the eluent. Further, the same coupling reaction was also tried under microwave conditions (100 W, 120 °C, 20–30 min), providing very good yields of the products 16(a–u) in comparison to the conventional heating method (Schemes 3 and 4).
8) as the eluent. Further, the same coupling reaction was also tried under microwave conditions (100 W, 120 °C, 20–30 min), providing very good yields of the products 16(a–u) in comparison to the conventional heating method (Schemes 3 and 4).
4.2 Antibacterial evaluation
Using agar well diffusion procedures, all of the synthesized compounds were tested for their antibacterial sensitivity in vitro against the test organisms, namely, the Gram-positive bacteria S. aureus and Gram-negative bacteria E. coli. Gentamicin was chosen as the standard drug. The synthesised compounds were first dissolved in DMSO. The zone of inhibition (ZI) was then evaluated using Mueller–Hinton agar plates. Initially, a sterilized Petri plate was filled with a 25 mL aliquot of the sterilized media. Once the media solidified, the microbial suspension was dispersed over the agar medium and a sterilized cork borer was used to create a well 6 mm in diameter. Next, for the ZI assessment, 80 μl of the test materials (100 μg mL−1) were placed into each well. Subsequently, the synthesized compounds were diluted at different concentrations (10, 20, 30, 40, 50, 60, 70, 80, and 100 μg mL−1) in order to determine the minimum inhibitory concentration (MIC) by assay. Next, the bacterial inhibitory efficacy was assessed for every fractionated entity in a 96-well plate experiment.26–284.3 Molecular docking studies
The docking computation of the synthesized compounds 16(a–u) was performed with AutoDock Tools version 4.2. The protein data bank (https://www.rcsb.org/) provided the crystal structures of the bacterial DNA gyrase proteins from E. coli and S. aureus. ChemDraw Ultra 12.0 was used to create the 3D structures of the synthesised ligands. During the molecular docking process, polar H-bonds were initially added, while confined ligands containing water molecules were removed, and the other default parameters were used. The docked complex of the ligand–receptor was visualised in two dimensions using PyMOL (https://www.pymol.org/) and BIOVIA Discovery Studio R2 2017. The best docking score was then chosen for additional evaluation of its antibacterial activities.294.4 ADME studies
The ADME and pharmacokinetic properties of all the synthesized compounds were studied on the free online server SwissADME (https://www.swissadme.ch/). Initially, chemical structures were drawn on Marvin to generate SMILES, which were then inserted directly on the webpage to initiate the prediction process. The resulting different pharmacokinetic properties and ADME parameters were obtained online and explained.23,30 The in silico ADMET predictions of the potent compound 16h were carried out using the web-based software ADMETlab 3.0 (https://admetlab3.scbdd.com/) using a methodology adopted from Rocha et al.31Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Rudra Narayan Mishra (RNM): conceptualization, methodology, investigation, formal analysis, data curation, software, writing review & editing; Mohammed Ansar Ahemad (MAA): methodology, formal analysis, data curation. Jasmine Panda (JP): writing original draft, conceptualization, methodology, resources, writing – review & editing. Sabita Nayak (SN): conceptualization, validation, supervision, project administration, funding acquisition, resources, writing – review & editing. Seetaram Mohapatra (SRM): supervision, resources, writing – review & editing, funding acquisition. Chita Ranjan Sahoo (CRS): antibacterial studies, resources, software.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
Author SN is thankful to DST ODISHA (2692/ST, Bhubaneswar, 16-7-2020) and LSRB New Delhi (LSRB/01/15001/M/LSRB-372/BTB/2020). Author RNM is thankful to DST ODISHA (Project no. ST-SCST-MISC-0011-2023) for financial support. Also, the author JP acknowledges INSPIRE Programme Division (Ref. DST/INSPIRE/03/2021/000846, New Delhi) for providing financial support in the form of INSPIRE FELLOWSHIP. Author SRM is thankful to SERB-SURE (Project No. SUR/2022/000257) for financial support in the form of research grant. And also thankful to the Centre of Excellence in Environment and Public Health by Higher Education Department, Government of Odisha (Grant no. 26913/HED/HE-PTC-WB-02-17 OHEPEE).References
- K. Allel, L. Day, A. Hamilton, L. Lin, L. Furuya-Kanamori, C. E. Moore, T. Van Boeckel, R. Laxminarayan and L. Yakob, Lancet Planet. Health, 2023, 7, e291–e303 CrossRef PubMed.
- (a) J. Baranwal, S. Kushwaha, S. Singh and A. Jyoti, Curr. Phys. Chem., 2023, 13, 2–19 CrossRef CAS; (b) J. Panda, B. P. Raiguru, S. Nayak, S. Mohapatra, S. Mohapatra, S. Rout, P. Mohanty, H. S. Biswal, D. Bhattacharya and C. R. Sahoo, New J. Chem., 2024, 48, 13609–13630 RSC; (c) B. Samanta, B. S. Panda, S. Mohapatra, S. Nayak, D. Bhattacharya and C. R. Sahoo, New J. Chem., 2024, 48, 4953–4959 RSC.
- (a) S. Renard, A. Olivier, P. Granger, P. Avenet, D. Graham, M. Sevrin, P. George and F. Besnard, J. Biol. Chem., 1999, 274, 13370–13374 CrossRef CAS PubMed; (b) M. Mishra, N. P. Mishra, B. P. Raiguru, T. Das, S. Mohapatra, S. Nayak, D. R. Mishra, J. Panda and D. K. Sahoo, ChemistrySelect, 2021, 6, 3570–3575 CrossRef CAS.
- (a) J. Panda, B. P. Raiguru, M. Mishra, S. Mohapatra and S. Nayak, ChemistrySelect, 2022, 7, e202103987 CrossRef CAS; (b) N. P. Mishra, S. Mohapatra, T. Das and S. Nayak, J. Heterocycl. Chem., 2022, 59, 2051–2075 CrossRef CAS.
- (a) H. Depoortere, B. Zivkovic, K. G. Lloyd, D. J. Sanger, G. Perrault, S. Z. Langer and G. Bartholini, J. Pharmacol. Exp. Ther., 1986, 237, 649–658 CAS; (b) S. S. Acharya, R. K. Sahoo, P. Mohanty, P. Panda and B. B. Parida, Chem. Pap., 2024, 78, 8107–8126 CrossRef CAS; (c) S. S. Acharya, P. Bhaumick, R. Kumar and L. H. Choudhury, ACS Omega, 2022, 7, 18660–18670 CrossRef CAS PubMed.
- N. Devi, A. K. Jana and V. Singh, Karbala Int. J. Mod. Sci., 2018, 4, 164–170 CrossRef.
- A. Kamal, J. S. Reddy, M. J. Ramaiah, D. Dastagiri, E. V. Bharathi, M. V. P. Sagar, S. Pushpavalli, P. Ray and M. Pal-Bhadra, MedChemComm, 2010, 1, 355–360 RSC.
- A. Deep, R. K. Bhatia, R. Kaur, S. Kumar, U. Kumar Jain, H. Singh, S. Batra, D. Kaushik and P. K. Deb, Curr. Top. Med. Chem., 2017, 17, 238–250 CrossRef CAS PubMed.
- S. Azimi, A. Zonouzi, O. Firuzi, A. Iraji, M. Saeedi, M. Mahdavi and N. Edraki, Eur. J. Med. Chem., 2017, 138, 729–737 CrossRef CAS PubMed.
- H. C. Kwong, C. C. Kumar, S. H. Mah, Y. L. Mah, T. S. Chia, C. K. Quah, G. K. Lim and S. Chandraju, Sci. Rep., 2019, 9, 926 CrossRef PubMed.
- B. K. R. Sanapalli, A. Ashames, D. K. Sigalapalli, A. B. Shaik, R. R. Bhandare and V. Yele, Antibiotics, 2022, 11, 1680, DOI:10.3390/antibiotics11121680.
- T. H. Al-Tel, R. A. Al-Qawasmeh and R. Zaarour, Eur. J. Med. Chem., 2011, 46, 1874–1881 CrossRef CAS PubMed.
- I. Althagafi and E. Abdel-Latif, Polycyclic Aromat. Compd., 2021, 42, 4487–4500 CrossRef.
- O. Ebenezer, P. Awolade, N. Koorbanally and P. Singh, Chem. Biol. Drug Des., 2020, 95, 162–173 CrossRef CAS PubMed.
- S. Kuthyala, M. K. Shankar and G. K. Nagaraja, ChemistrySelect, 2018, 3, 12894–12899 CrossRef CAS.
- A. Thakur, G. Pereira, C. Patel, V. Chauhan, R. K. Dhaked and A. Sharma, J. Mol. Struct., 2020, 1206, 1–13 CrossRef.
- N. P. Mishra, S. Mohapatra, C. R. Sahoo, B. P. Raiguru, S. Nayak, S. Jena and R. N. Padhy, J. Mol. Struct., 2021, 1246, 1–15 CrossRef.
- B. P. Raiguru, J. Panda, S. Mohapatra and S. Nayak, J. Mol. Struct., 2023, 1294, 136282 CrossRef CAS.
- B. P. Raiguru, S. Mohapatra, S. Nayak, D. Sahal, M. Yadav and B. N. Acharya, Eur. J. Med. Chem. Rep., 2022, 4, 100029 CAS.
- J. Panda, B. Brahma, S. Nayak, S. Mohapatra, B. P. Raiguru, S. P. Parida and K. Naithani, J. Heterocycl. Chem., 2025, 62, 99–121 CrossRef CAS.
- S. P. Parida, S. Mohapatra, S. Nayak, S. Mohapatra, J. Panda and C. R. Sahoo, ChemistrySelect, 2024, 9, e202402223 CrossRef.
- CCDC 2347891 (16e) contain the ESI† crystallographic data for this paper.
- (a) A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed; (b) O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed; (c) C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2012, 64, 4–17 CrossRef; (d) H. M. Faidallah and S. A. F. Rostom, Arch. Pharm., 2017, 350, 1–17 CrossRef PubMed.
- S. Rai, F. Ghous, S. Shukla, P. Sharma, P. Trivedi and A. Bishnoi, J. Mol. Struct., 2023, 1292, 136116 CrossRef CAS.
- M. R. Xavier, M. M. Marinho, M. S. S. Juliao, E. S. Marinho, F. W. Q. Almeida-Neto, K. K. A. deCastro, J. P. D. Hora, M. N. D. Rocha, A. C. H. Barreto, G. D. Saraiva, P. N. Bandeira, H. S. Santos, H. D. M. Coutinho and A. M. R. Teixeira, J. Mol. Struct., 2024, 1309, 138019 CrossRef CAS.
- C. R. Sahoo, S. Maharana, C. P. Mandhata, A. K. Bishoyi, S. K. Paidesetty and R. N. Padhy, Saudi J. Biol. Sci., 2020, 27, 1580–1586 CrossRef CAS PubMed.
- J. T. Poolman and A. S. Anderson, Expert Rev. Vaccines, 2018, 17, 607–618 CrossRef CAS PubMed.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS PubMed.
- C. R. Sahoo, S. K. Paidesetty, B. Dehury and R. N. Padhy, J. Biomol. Struct. Dyn., 2020, 38, 5419–5428 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, J. Chem. Inf. Model., 2014, 54, 3284–3301 CrossRef CAS PubMed.
- M. N. D. Rocha, D. R. Alves, M. M. Marinho, S. M. D. Morais, E. S. Marinho and J. Comput, Biophys. Chem., 2021, 20, 283–304 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2347891. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra09092f |
| This journal is © The Royal Society of Chemistry 2025 |