Open Access Article
Open Access ArticleA two-dimensional fluorescence and chemiluminescence orthogonal probe for discriminating and quantifying similar proteins†
Juan
Li
a,
Xiuyan
Zhao
a,
Yutao
Zhang
*a,
Yao
Lu
a,
Haoyun
Xue
a,
Dan
Li
a,
Qiang
Liu
a,
Chenxu
Yan
 a,
Weijie
Chi
a,
Weijie
Chi
 b,
Xingqing
Xiao
b,
Xingqing
Xiao
 *b,
Wei-Hong
Zhu
*b,
Wei-Hong
Zhu
 a and
Zhiqian
Guo
a and
Zhiqian
Guo
 *a
*a
aKey Laboratory for Advanced Materials, Institute of Fine Chemicals, Feringa Nobel Prize Scientist Joint Research Center, Frontiers Science Center for Materiobiology and Dynamic Chemistry, School of Chemistry and Molecular Engineering, Center of Photosensitive Chemicals Engineering, East China University of Science and Technology, Shanghai 200237, China. E-mail: zhangyutao@ecust.edu.cn; guozq@ecust.edu.cn
bDepartment of Chemistry, School of Chemistry and Chemical Engineering, Hainan University, Haikou City, Hainan Province 570228, China. E-mail: xqxiao@hainanu.edu.cn
First published on 7th January 2025
Abstract
Given that proteins with minor variations in amino acid sequences cause distinct functional outcomes, identifying and quantifying similar proteins is crucial, but remains a long-standing challenge. Herein, we present a two-dimensional orthogonal fluorescence and chemiluminescence design strategy for the probe DCM-SA, which is sequentially activated by albumin-mediated hydrolysis, exhibiting light-up fluorescence and photo-induced cycloaddition generating chemiluminescence, enabling orthogonal signal amplification for discrimination of subtle differences between similar proteins. By orthogonalizing these dual-mode signals, a two-dimensional work curve of fluorescence and chemiluminescence is established to distinguish and quantify similar proteins HSA and BSA. Importantly, the dual-mode signals of DCM-SA exhibit contrary incremental trends towards HSA and BSA. Molecular docking and femtosecond transient absorbance spectroscopy reveal that the lower KD value of DCM-SA with HSA and the longer excited-state lifetime of DCM-SA with BSA underlie the distinct dual-mode responses. Using two-dimensional orthogonal signals, for the first time, we precisely measure the HSA/BSA ratio in mixed serum. This method facilitates rapid blood source identification and trace HSA quantitation in human urine. Our two-dimensional orthogonal amplification approach offers a powerful tool for distinguishing and quantifying subtle differences among highly similar proteins, demonstrating great potential for both basic life science research and clinical applications.
Introduction
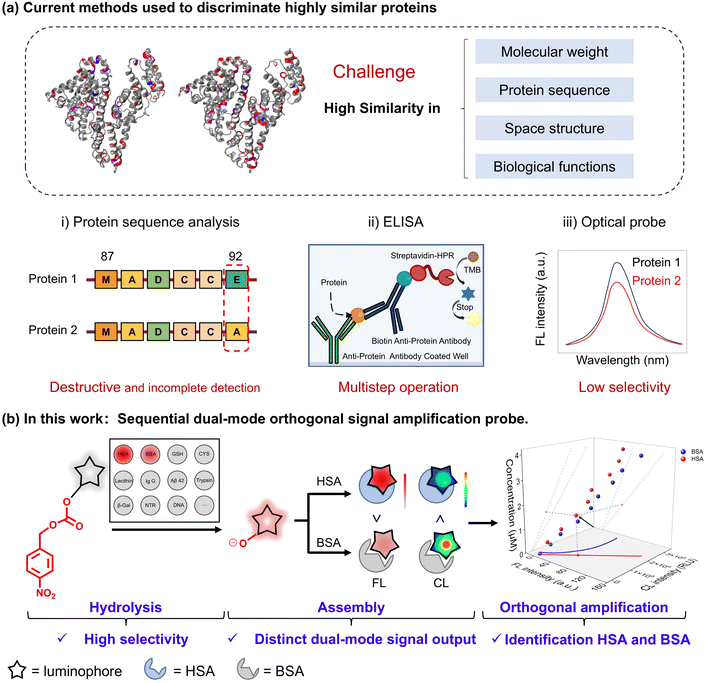
Serum albumin (SA), an essential blood protein, plays various roles including nutrient transportation, osmotic pressure regulation, and drug binding.1,2 Human serum albumin (HSA) and bovine serum albumin (BSA) have a similarity of 75.8% in terms of structure, chemical composition, and biological functions.3 However, unlike HSA, BSA cannot replenish lost fluids and restore blood volume, making their misuse potentially fatally harmful. Hence, distinguishing between HSA and BSA4 is crucial but highly challenging.The similar molecular weights of HSA and BSA render SDS-PAGE ineffective for differentiation.5 Amino acid sequencing like Edman degradation6 and mass spectrometry7 faces limitations in sequence analysis length, making it difficult to discriminate highly similar proteins.8 All these aforementioned procedures are destructive and require complex sample pre-handling and data analysis. Additionally, enzyme-linked immunosorbent assay9,10 (ELISA) through an antigen–antibody reaction suffers from the trade-off effect between sensitivity and selectivity to differentiate HSA and BSA. In contrast, optical probes offer rapid and convenient detection of target substances,11–19 but most of them merely rely on structural domains after protein binding, leading to lower selectivity when differentiating HSA from BSA (Fig. 1a). Therefore, it becomes an urgent demand to develop optical probes that could simultaneously identify and quantify HSA and BSA.
Proteins with highly similar structures can be distinguished via minor differences in amino acid sequences, electrostatic forces, van der Waals forces, charges, etc.13 This insight prompts us to develop a molecular probe strategy for detecting subtle differences in protein microenvironments. Towards this end, two crucial factors should be considered: first, the probe's ability to selectively bind to the protein; second, its ability to effectively sense and amplify the subtle interaction between the probe and albumin. Indeed, one-dimensional fluorescence signals alone have proven insufficient to simultaneously discriminate and quantify HSA and BSA. On the other hand, chemiluminescence is more easily influenced by environmental factors,20,21 such as polarity,22,23 hydrogen bonding,24–27 and pH,28–30 making it a valuable tool for sensing a protein's microenvironment. We hypothesize that by orthogonally coupling two-dimensional fluorescence and chemiluminescence, the subtle differences between HSA and BSA could be amplified. However, to date, such probes that meet the above requirements have yet to be reported.
Herein, we elaborately construct a two-dimensional probe DCM-SA that was sequentially triggered with hydrolysis-initiated fluorescence and photocycloaddition-induced chemiluminescence, allowing for orthogonal identification of similar proteins HSA and BSA. The probe DCM-SA is composed of two functional components: a 4-nitrobenzyl benzoate for specific recognition of SA species through albumin-catalyzed hydrolysis (Fig. 1b) and a luminophore, one of the dual-mode chemo-fluorophores, capable of luminophore–albumin assembly whose chemiluminescence could be activated subsequently upon light irradiation, enabling sensitivity to differentiate HSA and BSA. DCM-SA has a higher fluorescence signal for HSA compared to BSA, whereas the sequentially photoactivated chemiluminescence signal with HSA is lower than that of BSA. Molecule docking calculations and surface plasmon resonance (SPR) spectroscopy reveal that the tighter binding of the probe to albumins results in a higher fluorescence signal in response to HSA compared to BSA. Furthermore, femtosecond transient absorption spectroscopy (fs-TAS) suggests that the longer excited-state lifetime of the luminophore–BSA complex facilitates a higher chemiluminescence signal. The sequential two-dimensional fluorescence and chemiluminescence signals for HSA and BSA exhibit contrary incremental trends with concentration, thus effectively amplifying their subtle differences via orthogonal two-dimensional signals. The two-dimensional probe can effectively discriminate between human blood and bovine blood and accurately quantify the concentration of HSA in human urine. We provide a powerful tool for effectively detecting subtle differences in highly similar proteins via two-dimensional orthogonal response signals.
Results and discussion
Designing a two-dimensional probe for discrimination of HSA and BSA
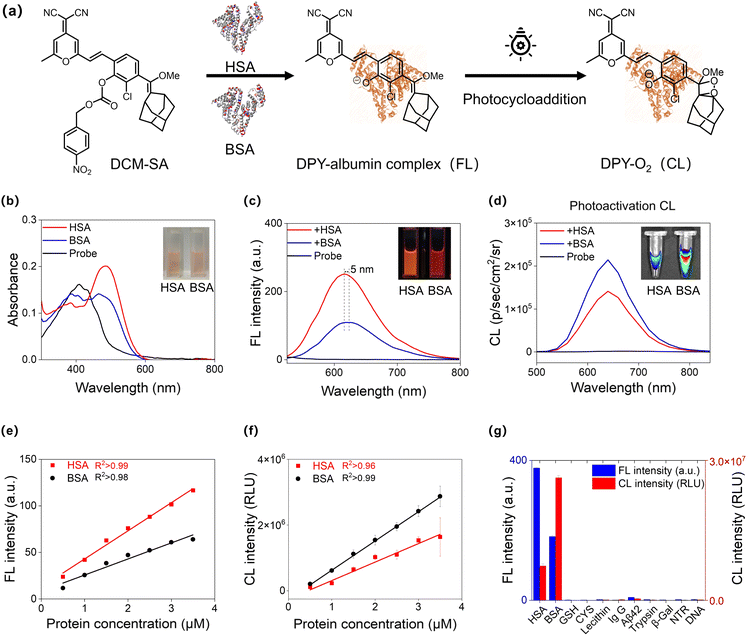
Toward this discrimination of HSA and BSA, we designed a fluorescence and chemiluminescence two-dimensional probe, DCM-SA, by incorporating the recognition unit 4-nitrobenzyl benzoate hydrolysable by SA and the photoactivatable chemo-fluorophore DPY (Fig. 2a). DPY was rationally designed to shorten the chemiluminescence half-life time based on our previous photoactivated chemo-fluorophore, thereby increasing the brightness and consequently improving the sensitivity of in vitro detection. In our strategy, two sequential31–33 triggers of SA and light were elaborately designed to guarantee specific recognition of SA through the light-up fluorescence signals from hydrolysis reactions, which released DPY. Subsequently, the deprotonated DPY exhibited a high binding affinity for albumins to form a DPY–albumin assembly, followed by the generation of markedly chemiluminescence signals upon light irradiation. Specifically, the ester bond between the chemo-fluorophore and the recognition unit was hydrolyzed, resulting in deprotonated DPY release accompanied by fluorescence emission. Then, the DPY–albumin complex was light-activated for in situ generation of high energy 1,2-dioxetane,34 an unstable cyclic peroxide, which decomposes to produce chemiluminescence.35–41 Above all, we presented this dual-mode probe named DCM-SA, which is capable of discriminating between HSA and BSA. All the compounds were characterized by 1H NMR, 13C NMR, and HRMS, as shown in Fig. S28–S36.†Investigating fluorescence and chemiluminescence two-dimensional responses towards HSA and BSA
Upon exposure to HSA/BSA, the absorption spectra of DCM-SA displayed a remarkable red-shift with maximum absorption at around 500 nm (Fig. 2b). The enhancement of fluorescence intensity depends on the incubation time with HSA/BSA, reaching a plateau at approximately 40 minutes (Fig. S1†). The fluorescence spectra of the probe at various pH levels (5–10) showed that DCM-SA responded to HSA/BSA under physiological pH conditions (Fig. S2 and S3†). At the same time, DCM-SA has good stability at room temperature (Fig. S4†). Notably, the fluorescence signal of DCM-SA in response to HSA was approximately 2.3 times higher than that of BSA (Fig. 2c and Table S1†). Unexpectedly, the chemiluminescence signal of DCM-SA in response to BSA was 1.5 times higher than that of HSA (Fig. 2d). We note that the inverse incremental ratio of these dual-mode signals significantly amplified the difference between HSA and BSA, making it possible to distinguish between the two albumins (Fig. S5–S7†). Moreover, we conducted a titration to investigate the fluorescence response of the probe DCM-SA to HSA/BSA. As the concentration of HSA and BSA increased (0–16 μM), the fluorescence intensity around 615 nm gradually increased and exhibited a linear correlation (Fig. 2e). Using the 3σ/slope method, the probe demonstrated a low limit of detection (LOD) in the fluorescence mode of 10 nM for HSA and 17 nM for BSA, respectively. Similarly, in the chemiluminescence mode, the LOD of the probe was calculated to be 8 nM for HSA and 2 nM (0.1 mg L−1) for BSA, respectively (Fig. 2f and S8, S9†). Additionally, after incubation with BSA, DCM-SA exhibited a red-shift of 5 nm in emission spectra compared to that of HSA, indicating that the microenvironment of the DPY–albumin complexes should be different. Based on these results of completely converse incremental trends of fluorescence and chemiluminescence intensities and obvious spectral shift, DCM-SA should be capable of distinguishing between HSA and BSA.Subsequently, we carried out the selectivity of DCM-SA to various amino acids, proteins, enzymes, and DNA (Fig. 2g and S10†). Clearly, DCM-SA was not influenced by potential competing species, showing exceptional selectivity towards HSA and BSA. Notably, a reference probe CF-Cl-NB with 4-nitrobenzyl as the response unit was designed (Fig. S11†), which did not show any response to HSA and BSA in either fluorescence or chemiluminescence mode. It was demonstrated that the excellent selectivity of DCM-SA was attributed to the specific response unit of 4-nitrobenzyl benzoate. Then, we verified the ester hydrolysis mechanism42 by HPLC analysis (Fig. S12†) and high-resolution mass spectrometry analysis (Fig. S13 and S14†). As shown in Fig. S12,† it unveiled the emergence of a peak after DCM-SA incubation with HSA/BSA, exhibiting a retention time of approximately 15 minutes after the response, aligning with DPY. Spectral response and HPLC tests indicated that it was the specific hydrolysis of the ester bond by albumin that conferred excellent selectivity of the probe towards HSA and BSA.
Revealing albumin microenvironment-mediated mechanism with molecular docking and femtosecond transient spectroscopy
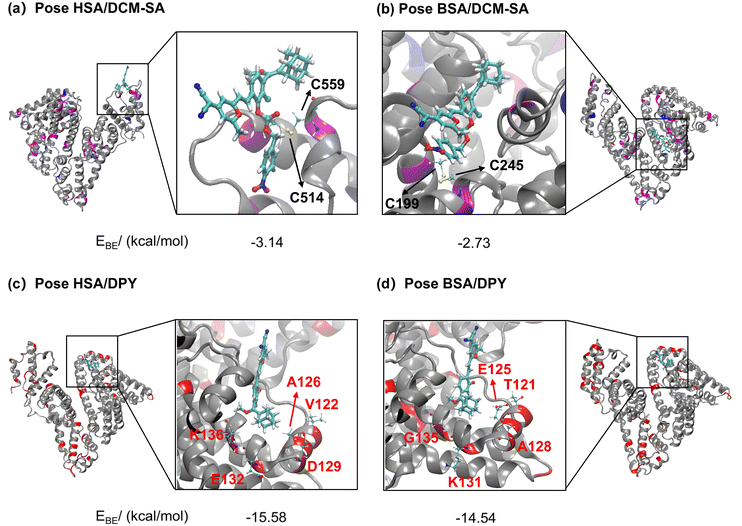
To gain insight into the binding disparities in the two-dimensional optical response, molecular docking simulations were conducted to model the interactions of DCM-SA and DPY with HSA and BSA, respectively (Fig. 3). The X-ray crystal structure of ligand-free HSA (ID: 1AO6) and BSA (4F5S) were collected from the Protein Data Bank. The top-ranked docking poses of HSA/DCM-SA, BSA/DCM-SA, HSA/DPY, and BSA/DPY complexes were calculated (Fig. S15–S19 and Tables S2 and S3†). The calculation results revealed a higher binding affinity between DCM-SA and HSA (−3.14 kcal mol−1) compared to BSA (−2.73 kcal mol−1), and the probes exhibited different binding sites in the two albumin species (Fig. 3a and b). Similarly, the DPY also displayed higher binding affinity to HSA (−15.58 kcal mol−1) than BSA (−14.54 kcal mol−1) (Fig. 3a and b).Then, we focused on exploring the microenvironment discrepancies of DPY at the binding sites in the two albumin species. Our emphasis was on residues with diverse properties (hydrophilicity, hydrophobicity, electronegativity) in the aligned sequences. It is obvious that the hydrophobic V122 and A126 tend to interact with DPY, making the HSA pocket nonpolar (Fig. 3c), while in the BSA pocket, the hydrophilic T121 and E125 preferentially associate with DPY, showing a somewhat strong polarity (Fig. 3d). ICT dyes have the property that the fluorescence wavelength red-shifts with increasing polarity of the solvent. Thus, the simulation results were well verified by fluorescence response tests (Fig. 2c), where the emission wavelength of DCM-SA was red-shifted by 5 nm upon incubation with BSA compared to HSA, indicating larger polarity within the BSA binding site. The above results suggested that differences in dye–albumin binding potentially underlie the discrepancies in the dual-mode optical response of DCM-SA to HSA and BSA.
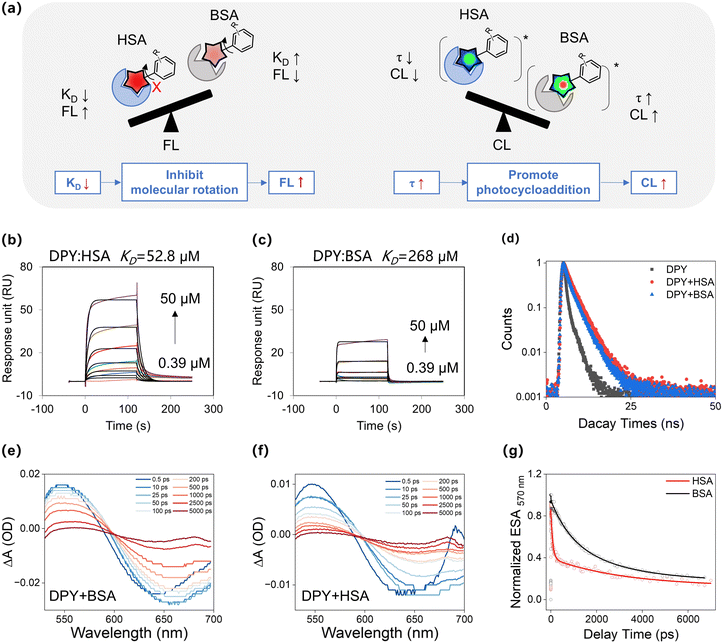
We further validated this molecular docking process through two consecutive tests (Fig. 4a): (i) investigating the impact of molecule rotation and its effect on the emission intensity of DPY. As expected, the fluorescence intensity of DPY has an increasing trend with the increase in the glycerol ratio (Fig. S20,†fG = 0–90%); (ii) surface plasmon resonance (SPR) assay was employed to measure the dissociation constant (KD) of DPY towards the two types of albumins. Consistent with molecular docking results, the results revealed that the KD value of HSA/DPY complexes (52.8 μM) was significantly lower than that of BSA/DPY complexes (Fig. 4b and c and Table S4†), confirming that the deprotonated DPY has a higher binding towards HSA. The data obtained from the double-logarithm plots of the protein quenching test and the steady-state lifetime of DPY/albumin complexes were consistent with the SPR results (Fig. 4d, S21, and S22 and Table S5†). These results collectively demonstrated that the stronger binding affinity of HSA with DPY inhibits molecular rotation, leading to a higher fluorescent response to HSA.
Subsequently, we investigated factors responsible for the higher bright chemiluminescence of the probe towards BSA (Fig. 4a). Femtosecond transient absorption spectroscopy (fs-TAS) is an important tool for studying photoreactions. We then investigated the lifetime (τ) of the excited state species of HSA/DPY complexes and BSA/DPY complexes (Fig. 4e and f). The excited state absorption at 570 nm was selected as a representative band to evaluate the excited state lifetimes of the two complexes. As shown in Fig. 4g, the excited state absorption signal of the HSA/DPY complexes at 570 nm decays rapidly with a lifetime of 0.96 ns. In contrast, the excited state absorption decay of the BSA/DPY complexes at 570 nm becomes significantly slow, with a lifetime of 2.36 ns. Long-lived excited state species can significantly promote photocycloaddition.43,44 Thus, the longer excited state lifetime of DPY in BSA solution promotes the photocyclization reaction of DPY, thereby exhibiting higher chemiluminescence. In short, molecular docking and femtosecond transient spectroscopy revealed that the lower KD values of DCM-SA with HSA along with the longer excited-state lifetime (τ) of DCM-SA with BSA were fundamental to the distinct differences in fluorescence and chemiluminescence responses to albumins in this microenvironment-mediated mechanism.
Establishing an orthogonal quantitative method for HSA and BSA
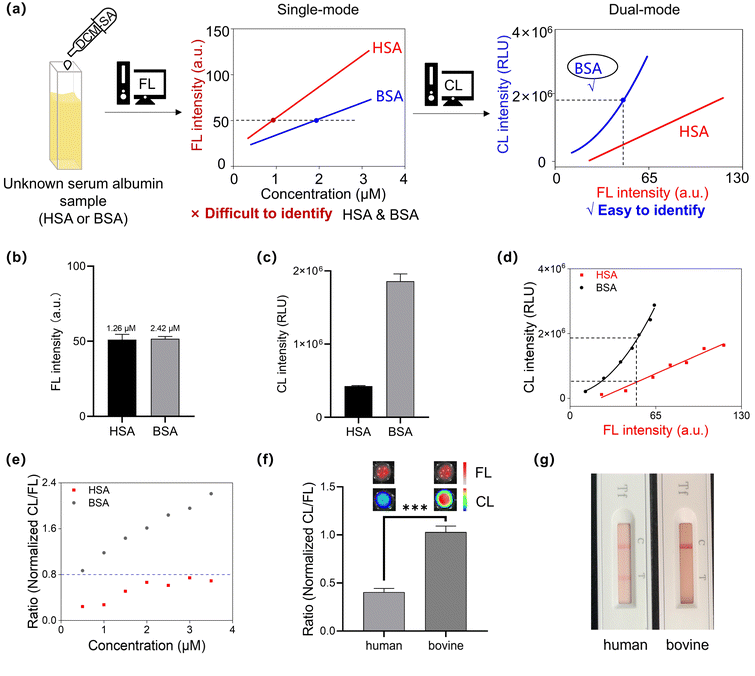
Indeed, one-dimensional and/or single-mode detection makes it difficult to determine the species and concentration of similar albumins simultaneously. For instance, one-dimensional 1H NMR often suffers from significant peak overlap in proteins, while two-dimensional NMR spectroscopy (such as C–H COSY spectra) provides orthogonal correlation shifts, enabling the analysis of subtle spatial conformational changes in proteins. Similarly, in our strategy, based on the linear one-dimensional relationship between fluorescence intensity and albumin concentration, we introduced a second-dimensional orthogonal relationship for chemiluminescence intensity. This approach allowed us to establish a hybrid fluorescence–chemiluminescence system that provides orthogonal signal outputs, thereby amplifying the subtle differences between similar albumins. As shown in Fig. 5a, relying solely on the fluorescence signal of DCM-SA was not sufficient to simultaneously identify and quantitate the albumin species. By incorporating chemiluminescence signals, we transited from one-dimensional to two-dimensional orthogonal localization, enabling exact determination of whether the sample was HSA or BSA. For instance, when an unidentified protein with a fluorescence intensity of 50 was detected, it could correspond to either 1.26 μM HSA or 2.42 μM BSA (Fig. 5a and b). Due to significant disparities in chemiluminescence exhibited by the probe with HSA and BSA (Fig. 5c), distinct orthogonal calibration curves for HSA and BSA were established for each albumin, enabling both differentiation and simultaneous quantitation (Fig. 5d). Additionally, DCM-SA can not only discriminate HSA and BSA, but also detect conformational changes in albumins resulting from temperature, heavy metals, and surfactants (Fig. S23†).Utilizing this two-dimensional orthogonal analysis method, we then used this probe to directly distinguish between human and bovine blood. As shown in Fig. 5e, we investigated the normalized CL/FL ratio of HSA with DCM-SA mixture and the BSA with DCM-SA mixture, respectively, and subsequently established the threshold as 0.8. Next, we employed the probe DCM-SA to measure the fluorescence and chemiluminescence signals of human and bovine blood (Fig. 5f). Notably, the ratio for human blood was 0.4, while that for bovine blood was 1.0. To validate the feasibility of the DCM-SA method, we used the anti-human transferrin colloidal gold test kit, a standard method in forensic investigations (Fig. 5g). The DCM-SA detection results were consistent with the colloidal gold test kit results, confirming the accuracy of DCM-SA in distinguishing between human and bovine blood (Fig. 5g and S25†).
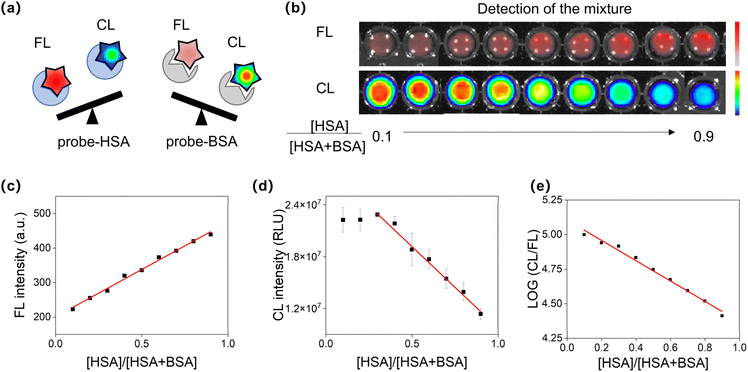
We then determined the precise proportion of HSA or BSA in a mixed serum (Fig. 6a). By maintaining a fixed total concentration of HSA and BSA, it was found that the fluorescence intensity of the mixture positively correlated with the percentage of HSA, while the chemiluminescence intensity exhibited a negative correlation (Fig. 6b–d). More importantly, a strong linear relationship (R2 = 0.98) was observed between the logarithm of the CL/FL ratio and the proportion of HSA (Fig. 6e), indicating that DCM-SA could precisely measure the percentage of the two proteins in the mixture. Overall, by utilizing the orthogonality of the chemiluminescence and fluorescence signals (Fig. 5d), we can not only rapidly distinguish human and bovine blood, but also accurately detect the ratio of HSA and BSA in a mixed serum.
Accurately diagnosing and quantifying HSA in clinical human urine
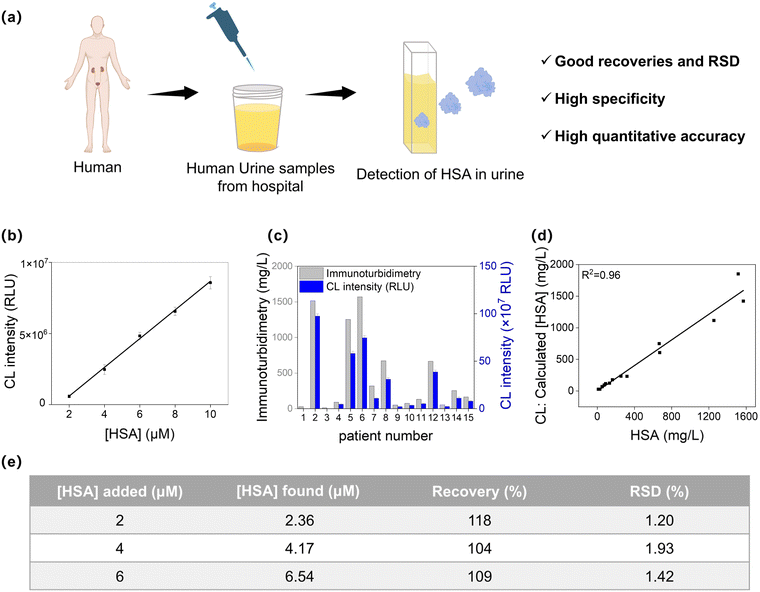
Urinary proteins comprise a variety of components, including albumin, microglobulin, macroglobulin, transferrin, and other proteins, with HSA making up around 60% of the total urinary protein.45–48 Particularly in the initial phases of glomerular diseases like early-stage diabetic nephropathy, early-stage hypertensive nephropathy, and early-stage chronic renal failure, HSA is the predominant protein detected in urine.49,50We evaluated the performance of DCM-SA for quantifying HSA concentration in human urine (Fig. 7a). The chemiluminescence intensity of DCM-SA displayed a good linear correlation with the HSA concentration (0–10 μM), and the LOD was calculated to be 0.4 μM (26 mg L−1, Fig. 7b and S26†). Additionally, quantitative analysis of HSA in human urine was performed by a standard recovery method with good recoveries and RSD (Fig. 7e). In fluorescence mode, DCM-SA had a similar behavior with a LOD of 0.2 μM (13 mg L−1, Fig. S27a†). Subsequently, DCM-SA was employed to measure HSA concentration in the urine samples of 15 patients, with results cross-validated by clinical immunoturbidimetry51 (Fig. 7c). Notably, curve-fitting analysis of the chemiluminescence data revealed a significant positive correlation between DCM-SA detection and immunoturbidimetry values, underscoring a remarkable linear relationship (CL: R2 = 0.96, Fig. 7d). Moreover, the detection accuracy in chemiluminescence mode was higher than that in fluorescence mode, especially at low concentrations (Fig. S27†). This result signified that DCM-SA was capable of accurately measuring HSA concentration in urine clinical samples. Thus, DCM-SA offered a convenient tool for assessing HSA in human urine and presented a novel alternative to conventional immunoassays.
Conclusions
In summary, we have developed a sequentially activated hydrolysis and photocycloaddition probe DCM-SA, enabling effective distinction and quantification of highly similar proteins. The probe DCM-SA exhibits contrary incremental trends of fluorescence and chemiluminescence signals for HSA and BSA, thereby orthogonally amplifying the subtle differences between these albumins. Specifically, via an albumin-catalyzed hydrolysis reaction, DCM-SA selectively detects serum albumins and assembles with them to form brightly fluorescent DPY–albumin complexes. Subsequently, under light irradiation, the DPY–albumin complexes undergo a cycloaddition reaction, producing chemiluminescence. By orthogonalizing these two-dimensional correlation signals, we amplify subtle differences between HSA and BSA. To our knowledge, for the first time, the two-dimensional optical probe accurately differentiates HSA and BSA. Molecular docking and surface plasmon resonance demonstrate that the stronger binding affinity of HSA with DPY (KD = 52.8 μM) inhibits molecular rotation, leading to higher fluorescence. Moreover, femtosecond transient absorbance spectroscopy reveals the longer excited state lifetime of the DPY–BSA complex, promoting photocycloaddition for higher chemiluminescence. We have successfully utilized the two-dimensional probe DCM-SA to identify the blood source, quantify the HSA/BSA mixed ratio, and detect trace HSA in human urine. Our two-dimensional orthogonal strategy offers a powerful tool for identifying and quantifying similar proteins, paving a new pathway for insight into minor variations in protein structures.Data availability
Experimental procedures and all relevant data are available in the ESI† and from the authors.Author contributions
All the experiments were conducted by J. L. and Y. Z. with the supervision of Z. G. and W. Z. All the computational work was conducted by X. X. and W. C. X. Z., H. X., and Q. L. repeated some experiments. Y. L., D. L., and C. Y. participated in the article discussions. All authors discussed the results and co-wrote the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Key Research and Development Program (2023YFA1802000), NSFC/China (22225805, 22308101, 22268016, 32394001, and 32121005), Shanghai Science and Technology Innovation Action Plan (No. 23J21901600), Shanghai Frontier Science Research Base of Optogenetic Techniques for Cell Metabolism (Shanghai Municipal Education Commission, grant 2021 Sci & Tech 03-28).Notes and references
- A. Jahanban-Esfahlan, A. Ostadrahimi, R. Jahanban-Esfahlan, L. Roufegarinejad, M. Tabibiazar and R. Amarowicz, Int. J. Biol. Macromol., 2019, 138, 602–617 CrossRef CAS.
- X. Li, S. Yu, Y. Lee, T. Guo, N. Kwon, D. Lee, S. C. Yeom, Y. Cho, G. Kim and J.-D. Huang, J. Am. Chem. Soc., 2018, 141, 1366–1372 CrossRef PubMed.
- A. Bujacz, Acta Crystallogr., Sect. D:Biol. Crystallogr., 2012, 68, 1278–1289 CrossRef CAS.
- C. Liu, W. Yang, Q. Gao, J. Du, H. Luo, Y. Liu and C. Yang, J. Lumin., 2018, 197, 193–199 CrossRef CAS.
- E. C. I. Veerman, R. J. F. Suppers, C. P. A. T. Klein and K. de Groot, Biomaterials, 1987, 8, 442–448 CrossRef CAS PubMed.
- P. Edman and G. Begg, Eur. J. Biochem., 1967, 1, 80–91 CrossRef CAS PubMed.
- K. F. Medzihradszky and R. J. Chalkley, Mass Spectrom. Rev., 2015, 34, 43–63 CrossRef CAS PubMed.
- K. Wang, S. Zhang, X. Zhou, X. Yang, X. Li, Y. Wang, P. Fan, Y. Xiao, W. Sun and P. Zhang, Nat. Methods, 2023, 21, 92–101 CrossRef PubMed.
- O. Weichenrieder and E. Izaurrald, Cell, 2018, 174, 5–6 CrossRef CAS.
- K. Zhang, C. Song, Q. Li, Y. Li, K. Yang and B. Jin, Hum. Vaccines, 2014, 6, 652–658 CrossRef PubMed.
- B. Liu, T. Lv, X. Zhao, M. Zhou, C. Song, C. Zeng, T. Qin and Z. Xu, Spectrochim. Acta, Part A, 2022, 264, 120306 CrossRef CAS PubMed.
- X.-J. Yan, Z. Li, H.-B. Liu, Z.-G. Wang, J. Fan, C.-Z. Xie, Q.-Z. Li and J.-Y. Xu, J. Photochem. Photobiol., A, 2022, 422, 113576 CrossRef CAS.
- A. Prah, J. Mavri and J. Stare, Phys. Chem. Chem. Phys., 2021, 23, 26459–26467 RSC.
- L. Zhang, Y. Zhang, W. Chi, C. Yan, Z. Zhao, X. Liu, W.-H. Zhu and Z. Guo, ACS Mater. Lett., 2022, 4, 1493–1502 CrossRef CAS.
- Y. Yu, X. Wang, X. Jia, Z. Feng, L. Zhang, H. Li, J. He, G. Shen and X. Ding, Adv. Sci., 2021, 8, 2102812 CrossRef CAS PubMed.
- X. Zhou, X. Su, D. Hu, Y. Li, L. Guo, W. Yuan, H. Yuan, L. Chen, M. Xu and S. Luo, ACS Appl. Mater. Interfaces, 2023, 15, 50916–50925 CrossRef CAS PubMed.
- Y. Ma, C. Yan, Z. Guo, G. Tan, D. Niu, Y. Li and W. H. Zhu, Angew. Chem., Int. Ed., 2020, 59, 21143–21150 CrossRef CAS PubMed.
- B. Liu, C. Zeng, D. Zheng, X. Zhao, C. Song, T. Qin and Z. Xu, Spectrochim. Acta, Part A, 2022, 274, 121081 CrossRef CAS PubMed.
- Y. Ke, D. Wei, Z. Lu, J. Cao, J. Liu and N. Fu, Sens. Actuators, B, 2023, 395, 134456 CrossRef CAS.
- H. Karatani, Bull. Chem. Soc. Jpn., 1987, 60, 2023–2029 CrossRef CAS.
- H. Umeda, K. Suda, D. Yokogawa, Y. Azumaya, N. Kitada, S. A. Maki, S. A. Kawashima, H. Mitsunuma, Y. Yamanashi and M. Kanai, Angew. Chem., Int. Ed., 2024, 63, e202405605 CrossRef CAS PubMed.
- N. Watanabe, H. K. Ijuin, H. Takatsuka and M. Matsumoto, Tetrahedron, 2021, 88, 132147 CrossRef CAS.
- M. Tanimura, N. Watanabe, H. K. Ijuin and M. Matsumoto, J. Org. Chem., 2010, 76, 902–908 CrossRef PubMed.
- N. Watanabe, H. Takatsuka, H. K. Ijuin, A. Wakatsuki and M. Matsumoto, Tetrahedron Lett., 2016, 57, 2558–2562 CrossRef CAS.
- N. Watanabe, H. Takatsuka, H. K. Ijuin and M. Matsumoto, Tetrahedron, 2020, 76, 131203 CrossRef CAS.
- J. Huang, P. Cheng, C. Xu, S. S. Liew, S. He, Y. Zhang and K. Pu, Angew. Chem., Int. Ed., 2022, 61, e202203235 CrossRef CAS PubMed.
- M. Matsumoto, T. Sakuma and N. Watanabe, Tetrahedron Lett., 2022, 43, 8955–8958 CrossRef.
- C. Chen, H. Gao, H. Ou, R. T. K. Kwok, Y. Tang, D. Zheng and D. Ding, J. Am. Chem. Soc., 2022, 144, 3429–3441 CrossRef CAS PubMed.
- T. Eilon-Shaffer, M. Roth-Konforti, A. Eldar-Boock, R. Satchi-Fainaro and D. Shabat, Org. Biomol. Chem., 2018, 16, 1708–1712 RSC.
- W. Adam, I. Bronstein, B. Edwards, T. Engel, D. Reinhardt, F. W. Schneider, A. V. Trofimov and R. F. Vasil’ev, J. Am. Chem. Soc., 1996, 118, 10400–10407 CrossRef CAS.
- Y. Ma, J. Shang, L. Liu, M. Li, X. Xu, H. Cao, L. Xu, W. Sun, G. Song and X.-B. Zhang, J. Am. Chem. Soc., 2023, 145, 17881–17891 CrossRef CAS PubMed.
- Y. Tao, C. Yan, D. Li, J. Dai, Y. Cheng, H. Li, W.-H. Zhu and Z. Guo, JACS Au, 2022, 2, 246–257 CrossRef CAS PubMed.
- Y. Zhang, C. Yan, C. Wang, Z. Guo, X. Liu and W. H. Zhu, Angew. Chem., Int. Ed., 2020, 59, 9059–9066 CrossRef CAS PubMed.
- S. Gutkin, R. Tannous, Q. Jaber, M. Fridman and D. Shabat, Chem. Sci., 2023, 14, 6953–6962 RSC.
- O. Green, T. Eilon, N. Hananya, S. Gutkin, C. R. Bauer and D. Shabat, ACS Cent. Sci., 2017, 3, 349–358 CrossRef CAS PubMed.
- H. N. Kagalwala, J. Gerberich, C. J. Smith, R. P. Mason and A. R. Lippert, Angew. Chem., Int. Ed., 2022, 61, e202115704 CrossRef CAS PubMed.
- H. N. Kagalwala, R. T. Reeves and A. R. Lippert, Curr. Opin. Chem. Biol., 2022, 68, 102134 CrossRef CAS PubMed.
- M. Zhao, Y. Lu, Y. Zhang, H. Xue and Z. Guo, Chin. Chem. Lett., 2024 DOI:10.1016/j.cclet.2024.110105.
- Y. Lu, Y. Zhang, X. Wu, R. Pu, C. Yan, W. Liu, X. Liu, Z. Guo and W.-H. Zhu, Chem. Sci., 2024, 15, 12431–12441 RSC.
- C. Chen, H. Gao, H. Ou, R. T. K. Kwok, Y. Tang, D. Zheng and D. Ding, J. Am. Chem. Soc., 2022, 144, 3429–3441 CrossRef CAS.
- L. Chen, K. Sun, D. Hu, X. Su, L. Guo, J. Yin, Y. Pei, Y. Fan, Q. Liu and M. Xu, Angew. Chem., Int. Ed., 2023, 62, e202218670 CrossRef CAS PubMed.
- K. Kono, Y. Fukuchi, H. Okawa, K.-i. Nunoya, H. Imawaka, H. Watanabe and T. Maruyama, Mol. Pharm., 2019, 16, 4131–4138 CrossRef CAS.
- S. Poplata, A. Tröster, Y.-Q. Zou and T. Bach, Chem. Rev., 2016, 116, 9748–9815 Search PubMed.
- M. J. Genzink, M. D. Rossler, H. Recendiz and T. P. Yoon, J. Am. Chem. Soc., 2023, 145, 19182–19188 Search PubMed.
- S. Decramer, A. G. de Peredo, B. Breuil, H. Mischak, B. Monsarrat, J.-L. Bascands and J. P. Schanstra, Mol. Cell. Proteomics, 2008, 7, 1850–1862 CrossRef CAS PubMed.
- S. Guan, Q. Fu, D. Wang, Y. Han, N. Cao, M. Zhang, H. Shen, R. Yang, B. He and M. Tao, ACS Sens., 2022, 7, 3481–3490 CrossRef CAS PubMed.
- Z. Luo, T. Lv, K. Zhu, Y. Li, L. Wang, J. J. Gooding, G. Liu and B. Liu, Angew. Chem., Int. Ed., 2020, 59, 3131–3136 CrossRef CAS PubMed.
- S. Sarkar, A. Shil, Y. L. Jung, S. Singha and K. H. Ahn, ACS Sens., 2022, 7, 3790–3799 CrossRef CAS PubMed.
- B. Liu, K. Zhu and T. Lü, Chin. J. Org. Chem., 2019, 39, 2786–2795 CrossRef.
- S. Park and H.-J. Kim, Sens. Actuators, B, 2012, 168, 376–380 CrossRef CAS.
- R. Flores, Anal. Biochem., 1978, 88, 605–611 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc07714h |
| This journal is © The Royal Society of Chemistry 2025 |