Open Access Article
Open Access ArticleOrthologous mammalian A3A-mediated single-nucleotide resolution sequencing of DNA epigenetic modification 5-hydroxymethylcytosine†
Xia
Guo‡
abc,
Jianyuan
Wu‡
d,
Tong-Tong
Ji
c,
Min
Wang
a,
Shan
Zhang
c,
Jun
Xiong
a,
Fang-Yin
Gang
a,
Wei
Liu
a,
Yao-Hua
Gu
ae,
Yu
Liu
*af,
Neng-Bin
Xie
*a and
Bi-Feng
Yuan
 *abc
*abc
aDepartment of Occupational and Environmental Health, School of Public Health, Wuhan University, Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, Wuhan 430071, China. E-mail: bfyuan@whu.edu.cn; nengbinxie@whu.edu.cn; liuyu97@whu.edu.cn
bResearch Center of Public Health, Renmin Hospital of Wuhan University, Wuhan 430060, China
cCollege of Chemistry and Molecular Sciences, Hubei Key Laboratory of Biomass Resource Chemistry and Environmental Biotechnology, Wuhan University, Wuhan 430072, China
dClinical Trial Center, Zhongnan Hospital of Wuhan University, Wuhan 430071, China
eSchool of Nursing, Wuhan University, Wuhan 430071, China
fHubei Key Laboratory of Tumor Biological Behaviors, Cancer Clinical Study Center, Zhongnan Hospital of Wuhan University, Wuhan 430071, China
First published on 28th January 2025
Abstract
Epigenetic modifications in genomes play a crucial role in regulating gene expression in mammals. Among these modifications, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are recognized as the fifth and sixth nucleobases in genomes, respectively, and are the two most significant epigenetic marks in mammals. 5hmC serves as both an intermediate in active DNA demethylation and a stable epigenetic modification involved in various biological processes. Analyzing the location of 5hmC is essential for understanding its functions. In this study, we introduce an orthologous mammalian A3A-mediated sequencing (OMA-seq) method for the quantitative detection of 5hmC in genomic DNA at single-nucleotide resolution. OMA-seq relies on the deamination properties of two naturally occurring mammalian A3A proteins: green monkey A3A (gmA3A) and dog A3A (dogA3A). The combined use of gmA3A and dogA3A effectively deaminates cytosine (C) and 5mC, but not 5hmC. As a result, the original C and 5mC in DNA are deaminated and read as thymine (T) during sequencing, while the original 5hmC remains unchanged and is read as C. Consequently, the remaining C in the sequence indicates the presence of original 5hmC. Using OMA-seq, we successfully quantified 5hmC in genomic DNA from lung cancer tissue and corresponding normal tissue. OMA-seq enables accurate and quantitative mapping of 5hmC at single-nucleotide resolution, utilizing a pioneering single-step deamination protocol that leverages the high specificity of natural deaminases. This approach eliminates the need for bisulfite conversion, DNA glycosylation, chemical oxidation, or screening of engineered protein variants, thereby streamlining the analysis of 5hmC. The utilization of orthologous enzymes for 5hmC detection expands the toolkit for epigenetic research, enabling the precise mapping of modified nucleosides and uncovering new insights into epigenetic regulation.
Introduction
Epigenetic modifications in nucleic acids are critical regulators of gene expression in mammals.1 5-Methylcytosine (5mC) represents the first and most extensively studied modified nucleobase in DNA, playing a pivotal role in fundamental biological processes including gene expression regulation and normal developmental progression.2 The discovery of 5-hydroxymethylcytosine (5hmC) in mammalian genomes marked a significant milestone in epigenomics research.3,4 5hmC is generated from 5mC through an active DNA demethylation process catalyzed by the ten-eleven translocation (TET) family of dioxygenases.5–8 Beyond its role as an intermediate in DNA demethylation, 5hmC is now recognized as a distinct epigenetic mark and functionally significant nucleobase within mammalian genomic landscapes.9 Emerging research increasingly highlights the multifaceted roles of 5hmC in diverse biological processes, including tumorigenesis and embryonic development.105hmC is generally linked to active gene expression and may indicate transcription that is dynamically activated.10 The distribution of 5hmC is specific to different tissues, with a higher prevalence in genes that promote tissue differentiation and in tissue-specific transcription factors.11 Additionally, 5hmC plays a crucial role in sustaining a pluripotent state in cells as well as in cell differentiation.12 In the context of cancer, both a widespread reduction of 5hmC and localized increases in genes were observed during tumour progression.13 Therefore, analyzing 5hmC offers valuable insights into epigenetic activation during the course of tumour development.
Some methods have been developed to detect 5hmC in genomic DNA, including thin layer chromatography detection,3,4 and liquid chromatography coupled with mass spectrometry analysis.14–21 Typically, these approaches involve digesting genomic DNA into nucleosides or nucleotides for qualitative and quantitative 5hmC detection.22 Understanding the functional significance of 5hmC in DNA requires precise location analysis within genomic landscapes.23 Some techniques have emerged for mapping 5hmC in DNA. Hydroxymethylated DNA immunoprecipitation (hMeDIP) using specific antibodies and affinity-based purification and sequencing methods enable whole-genome 5hmC mapping.24,25 However, these approaches are limited by their resolution, which depends on the genomic DNA fragment size, preventing identification of exact 5hmC sites.26 Oxidative bisulfite sequencing (oxBS-seq)27 and TET-assisted bisulfite sequencing (TAB-seq)28 were introduced to achieve single-nucleotide resolution 5hmC detection. A limitation of these methods is the use of bisulfite, which can cause significant DNA degradation, potentially destroying up to 99.9% of input genetic material.29 Single-molecule real-time (SMRT) sequencing offers another 5hmC detection avenue,30 though it currently suffers from a high false-positive rate in modification mapping.31 More recent innovations include TET-assisted pyridine borane sequencing (TAPS), β-glucosyltransferase blocking TET-assisted pyridine borane sequencing (TAPSβ), and chemical-assisted pyridine borane sequencing (CAPS), which aim to map 5hmC at single-base resolution.32,33 Nevertheless, these methods face challenges, as the dihydrouracil (DHU)-containing DNA produced can compromise amplification efficiency, potentially introducing bias in genome-wide 5hmC mapping.34
More recently, enzyme-assisted methods in mapping modified nucleobases have been developed.35–40 The A3A (apolipoprotein B mRNA-editing catalytic polypeptide-like 3A) protein, present across most placental mammals, enables deamination of cytosines to uracil in DNA.41,42 It has been reported that human A3A (hA3A) protein can efficiently deaminate cytosine (C), 5mC, and 5hmC in DNA, but shows no deamination activity toward glycosylated 5hmC (β-glucosyl-5-hydroxymethyl-2′-deoxycytidine, 5gmC).43–46 Using this property of hA3A, we and others have reported engineered deaminase-mediated sequencing (EDM-seq),47 single-step deamination sequencing (SSD-seq),48 and A3A-coupled epigenetic sequencing (ACE-seq)49 for mapping 5hmC in DNA. In ACE-seq, 5hmC is first glycosylated to produce 5gmC by using β-glucosyltransferase (β-GT). hA3A treatment leads to the conversion of C and 5mC in DNA to uracil (U) and thymine (T), respectively. Both U and T are read as T, while 5gmC resists deamination and is read as C during sequencing. These methods, however, require the pre-treatment of DNA with β-GT to glycosylate 5hmC or demand laborious screening of engineered hA3A variants.
Previous studies have demonstrated that orthologous mammalian A3A cytidine deaminases exhibit varying degrees of deamination activity toward C and 5mC.41,50,51 Apart from hA3A, the deaminase activity of other orthologous mammalian A3A proteins toward 5hmC remained largely unexplored. In this study, we comprehensively examined the deamination activities of six orthologous mammalian A3A proteins on C, 5mC, and 5hmC, revealing two distinct types of mammalian A3A proteins with specific sequence preferences. Green monkey A3A (gmA3A, from Chlorocebus aethiops) demonstrated complete deamination of C and 5mC but showed no deamination activity toward 5hmC at YC (Y = C/T) sites. Conversely, dog A3A (dogA3A, from Canis lupus familiaris) could fully deaminate C and 5mC but exhibited no deamination activity toward 5hmC at RC (R = A/G) sites. Leveraging these unique deamination properties of gmA3A and dogA3A, we proposed an orthologous mammalian A3A-mediated sequencing (OMA-seq) method for single-nucleotide resolution and quantitative analysis of 5hmC. OMA-seq approach introduces novel, naturally occurring enzymes for precise 5hmC mapping and quantification.
Experimental section
Chemicals and reagents
The 24-mer cytosine-containing DNA, 24-mer 5mC-containing DNA, as well as the 24-mer 5hmC-containing DNA were synthesized by Takara Biotechnology Co., Ltd (Dalian, China). Other single-stranded DNAs were purchased from Tianyi Huayu Gene Technology Co., Ltd (Wuhan, China). The sequences of these oligonucleotides are provided in Table S1.† 2′-Deoxycytidine (dC), 2′-deoxyguanosine (dG), 2′-deoxyadenosine (dA), 2′-deoxythymidine (dT), and the 2′-deoxynucleoside 5′-triphosphates, including dATP, dCTP, dGTP, and TTP, as well as venom phosphodiesterase I, were purchased from Sigma-Aldrich (Beijing, China). 5-Methyl-2′-deoxycytidine-5′-triphosphate (5mdCTP) and 5-hydroxymethyl-2′-deoxycytidine-5′-triphosphate (5hmdCTP) were obtained from TriLink BioTechnologies (San Diego, California, USA). DNase I, S1 nuclease, and alkaline phosphatase were purchased from Takara Biotechnology Co., Ltd. Lung cancer tissue and the matched adjacent normal tissue samples were collected from Zhongnan Hospital of Wuhan University. All experiments were conducted in compliance with the guidelines and regulations established by the Ethics Committee of Wuhan University.In vitro expression and purification of orthologous mammalian A3A proteins
The sequences for orthologous mammalian A3A proteins are obtained from the NCBI database and are listed in Table S2.† For the expression and purification of the human A3A (hA3A) protein, the coding sequence was incorporated into the pET-41a(+) plasmid, which was synthesized de novo by TsingKe Co., Ltd (Beijing, China). This plasmid includes a human rhinovirus 3C protease (HRV 3C) site between the glutathione S-transferase (GST) tag and the hA3A protein sequence. The resulting plasmid, pET-41a(+)-hA3A, was transformed into the E. coli BL21(DE3) pLysS strain (Sangon). The detailed procedures for the in vitro expression and purification of hA3A could be found in the ESI.† The purity of the protein was assessed by SDS-PAGE, and was stored at −80 °C after the addition of 25% glycerol. The concentration of the purified protein was quantified using the BCA protein assay kit (Beyotime). The expression and purification of other mammalian A3A proteins, including cowA3A, pandaA3A, porpoiseA3A, gmA3A, and dogA3A, were conducted in a similar manner to that of hA3A.Preparation of DNA with C, 5mC, and 5hmC modifications
The 216-bp double-stranded DNA (dsDNA) substrates, including DNA-C, DNA-5mC, and DNA-5hmC, were prepared for the sequencing method development. Detailed sequences are provided in Table S3.† Briefly, a 228-bp DNA (Table S3†) was synthesized and inserted into the pET-41a plasmid (pET41a-DNA-C) using SpeI and XhoI cloning sites (TsingKe). DNA-C was generated by PCR amplification with 2 ng of pET41a-DNA-C serving as the template. The PCR amplification was conducted in a 50 μL reaction mixture containing 2.5 U of Taq DNA polymerase (Accurate), 1× reaction buffer, 0.2 mM each of dATP, dGTP, TTP, and dCTP, 0.4 μM forward primer (5′-AGTGACGCTGAGCTTGACGTCGCGC-3′), and 0.4 μM reverse primer (5′-CCAACATTCCACTAACAATTACTCTCT-3′). The PCR program included 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, followed by 72 °C for 5 min. DNA-5mC and DNA-5hmC were prepared similarly, with dCTP substituted by 5mdCTP and 5hmdCTP, respectively. The PCR products were verified by agarose gel electrophoresis and purified using KAPA Pure beads (Roche).Deamination assay for dsDNA carrying various modifications
DNA-C, DNA-5mC, and DNA-5hmC were used to evaluate the deaminase activity of orthologous mammalian A3A proteins toward C, 5mC, and 5hmC. Typically, 60 ng of the dsDNA substrate was denatured by adding 2 μL of DMSO and heating at 95 °C for 10 min, followed by immediate cooling in ice water. The deamination reaction was conducted in a 20 μL solution containing the denatured DNA, 10 μM of the orthologous mammalian A3A protein, 20 mM MES (pH 6.5), and 0.1% Triton X-100. The reaction proceeded with a temperature gradient of 4 °C for 5 min, followed by a linear ramp from 4 to 37 °C at a rate of +0.1 °C every 13 s, then held at 37 °C for 2 h, and finally a linear ramp from 37 to 50 °C at a rate of +0.1 °C every 13 s. The reaction was terminated by heating at 95 °C for 15 min. Deaminase-treated DNA was subjected to PCR amplification. The PCR amplification was conducted in a 50 μL reaction mixture containing 1.25 U of EpiMark Hot Start Taq DNA polymerase (New England Biolabs), 1× reaction buffer, 0.2 mM each of dATP, dGTP, TTP, and dCTP, 0.4 μM forward primer (5′-AGTGATGTTGAGTTTGATGTTGTGT-3′) and 0.4 μM reverse primer (5′-CCAACATTCCACTAACAATTACTCTCT-3′). The PCR program included 95 °C for 5 min, 25 cycles of 95 °C for 30 s, 55 °C for 30 s, 68 °C for 30 s, followed by 68 °C for 5 min. The resulting PCR products were subjected to Sanger sequencing and colony sequencing (TsingKe). Colony sequencing was performed according to previously reported method,52,53 with detailed procedures available in the ESI.†Deamination assay for ssDNA with various modifications
The 24-mer cytosine-containing DNA, 24-mer 5mC-containing DNA, and the 24-mer 5hmC-containing DNA were used to evaluate the deaminase activity of orthologous mammalian A3A proteins toward C, 5mC, and 5hmC. The deamination reaction was conducted in a 20 μL solution containing 10 μM of the orthologous mammalian A3A protein, 20 mM MES (pH 6.5), 0.1% Triton X-100, and 60 ng of the oligonucleotide at 37 °C for 2 h. The reaction was terminated by heating at 95 °C for 15 min. The deamination rates of the orthologous mammalian A3A proteins were determined using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.Enzymatic digestion of DNA
DNA was enzymatically digested according to the previously described method,54 and the detailed procedures are available in the ESI.†LC-MS/MS analysis
The analysis of nucleosides was conducted according to previous studies.55–58 The measurements were performed using an LC-MS/MS system, which included a Shimadzu 8045 mass spectrometer (Kyoto, Japan) with a Turbo Ionspray electrospray ionization source, coupled with a Shimadzu LC-30AD UPLC system. The nucleosides were monitored using multiple reaction monitoring (MRM) in positive-ion mode. The optimized MRM parameters are provided in Table S4.† The detailed analytical procedures are available in the ESI.†Quantitative analysis of 5hmC in specific loci of lung tissue DNA
Genomic DNA from lung cancer tissue and matched adjacent normal tissue were extracted using a tissue DNA kit (Omega Bio-Tek Inc., Norcross, GA, USA). The genomic DNA was fragmented to an average size of 300–500 bp using an Ultrasonic Homogenizer JY92-IIN (Scientz). For OMA-seq, 60 ng of fragmented DNA was denatured by heating and treated with 10 μM of orthologous mammalian A3A protein. For ACE-seq, 1 mg of fragmented DNA was treated with β-GT (New England Biolabs) at 37 °C for 2 h, followed by recovery using KAPA Pure beads (Roche). Subsequently, 60 ng of the treated DNA was denatured by heating and then treated with 10 μM of hA3A protein. The deaminase-treated DNA was amplified using EpiMark Hot Start Taq DNA polymerase (New England Biolabs) with site-specific primers (Table S5†) for the individual test sites. The PCR products were subjected to Sanger sequencing, and the quantification of 5hmC levels was achieved by calculating the peak heights of C and T at the test sites.Results and discussion
Characteristics of different orthologous mammalian A3A proteins
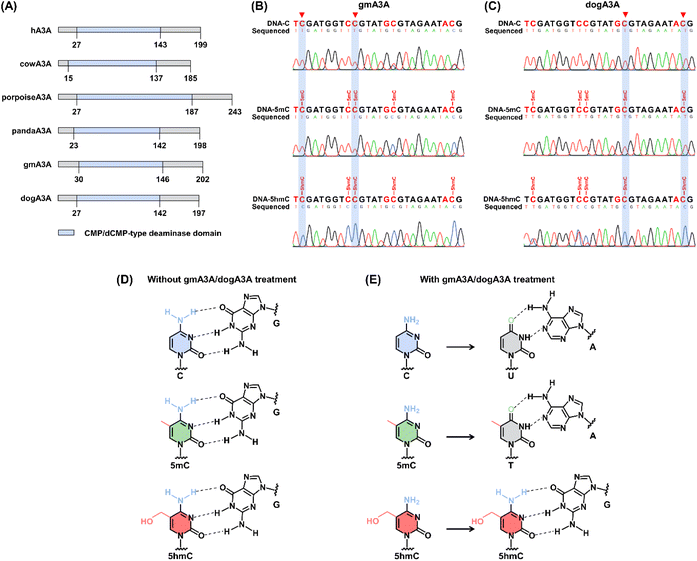
A3A is a universal deaminase found in most placental mammals.41 However, only a few studies have reported the deamination activity of orthologous mammalian A3A proteins toward C and 5mC. Except for hA3A, the deamination characteristics of other orthologous mammalian A3A members toward 5mC and 5hmC remains inefficiently explored. In the current study, we expressed six orthologous mammalian A3A proteins in vitro, including human A3A (hA3A), cow A3A (cowA3A), panda A3A (pandaA3A), porpoise A3A (porpoiseA3A), green monkey A3A (gmA3A), and dog A3A (dogA3A). We carefully assessed their deamination activity toward C, 5mC, and 5hmC, as well as their substrate preferences. A schematic illustration of these six orthologous mammalian A3A proteins is shown in Fig. 1A. Alignment of these proteins revealed approximately 41% amino acid identity (Fig. S1†).The deamination activity of hA3A toward cytosine has been reported to be influenced by the adjacent 5′ nucleobase.43,50 We designed three dsDNA substrates (DNA-C, DNA-5mC, and DNA-5hmC) to further evaluate the readouts of C, 5mC, and 5hmC in different sequence contexts (TC, CC, GC, and AC). Without deamination, C, 5mC, and 5hmC base pair with guanine (G) and are read as C in sequencing. However, if C, 5mC, and 5hmC are deaminated, they are converted into uracil (U), thymine (T), and 5-hydroxyuracil (5hmU), respectively, which base pair with adenine (A) and are read as T in sequencing. The dsDNA was denatured to ssDNA and then treated with hA3A. The resulting DNA was amplified by PCR and subjected to Sanger sequencing. The results showed that all cytosines in DNA-C and all 5mC in DNA-5mC were fully deaminated and read as thymine (T) after hA3A treatment (Fig. S2†). In contrast, the 5hmC in DNA-5hmC was completely read as T in YC (Y = C/T) sites but only partially read as T in RC (R = A/G) sites. This indicates that hA3A fully deaminates 5hmC at YC sites while partially deaminating 5hmC at RC sites (Fig. S2†).
We then evaluated the characteristics of the other five orthologous mammalian A3A proteins. For cowA3A, the results showed that cytosines in DNA-C and 5mC in DNA-5mC were partially deaminated (Fig. S3†), indicating relatively low deamination activity. For porpoiseA3A, cytosines in DNA-C at RC sites and 5mC at CC sites were also partially deaminated (Fig. S4†), suggesting similarly low activity. In the case of pandaA3A, cytosines in DNA-C at RC sites were partially deaminated, while 5mC in DNA-5mC at RC sites was not deaminated, and at CC sites, it was partially deaminated (Fig. S5†). None of these three A3A proteins exhibited deamination activity toward 5hmC. These results suggest that hA3A, cowA3A, porpoiseA3A, and pandaA3A are unsuitable for method development.
For gmA3A, the results showed that all cytosines in DNA-C and all 5mC in DNA-5mC were fully deaminated and read as T, while all 5hmC in DNA-5hmC resisted deamination and was read as C at YC sites (Fig. 1B and S6†). Similarly, after treatment with dogA3A, all cytosines in DNA-C and all 5mC in DNA-5mC were read as T, while all 5hmC in DNA-5hmC was read as C at RC sites (Fig. 1C and S7†). The combined use of gmA3A and dogA3A may enable the distinction of 5hmC from C and 5mC at all sites, which encourages us to employ them in developing a method to detect 5hmC at single-base resolution (Fig. 1D and E).
OMA-seq
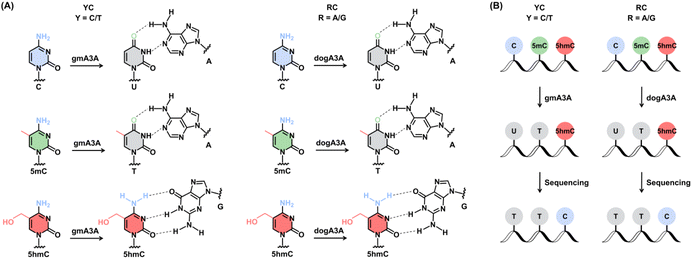
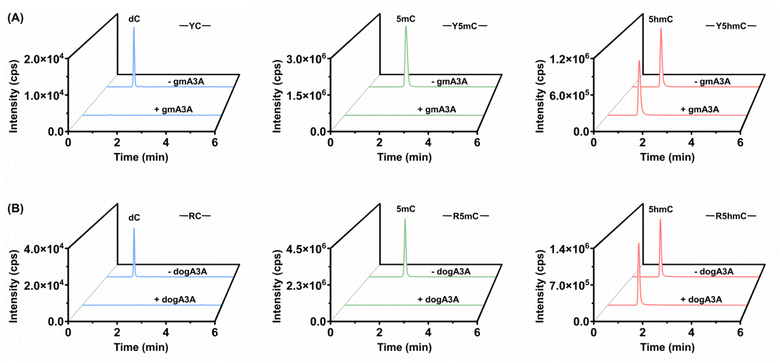
Leveraging the complementary deamination properties of gmA3A and dogA3A toward C, 5mC, and 5hmC in different sequence contexts, we developed an orthologous mammalian A3A-mediated sequencing (OMA-seq) method for single-nucleotide resolution and quantitative detection of 5hmC (Fig. 2). At YC (Y = C/T) sites, gmA3A fully deaminates C and 5mC to form U and T, which base pair with A and are read as T, while showing no deamination activity toward 5hmC, which remains paired with G and is read as C in sequencing (Fig. 2A). At RC (R = A/G) sites, dogA3A similarly fully deaminates C and 5mC to U and T, which base pair with A and are read as T, while also showing no deamination activity toward 5hmC, which remains paired with G and is read as C (Fig. 2A). The detection of 5hmC in various sequence contexts, including TC, CC, GC, and AC sites, is achieved through the combined use of gmA3A and dogA3A. In OMA-seq, gmA3A detects 5hmC at TC and CC sites, while dogA3A detects it at GC and AC sites, with C and 5mC read as T, and 5hmC read as C (Fig. 2B).To gain further insight into the deamination activity of gmA3A and dogA3A, we conducted LC-MS/MS analysis using various substrates. This analysis evaluated the deamination efficiency of gmA3A and dogA3A toward C, 5mC, and 5hmC. Based on the property of gmA3A for deaminating C, 5mC, and 5hmC, we treated three different DNA mixtures (TC-C and CC-C; TC-5mC and CC-5mC; TC-5hmC and CC-5hmC) with gmA3A, followed by LC-MS/MS analysis. The results showed that the signals for dC and 5mC were undetectable, while the signal intensity of 5hmC remained nearly intact after treatment with gmA3A (Fig. 3A). Similarly, we treated another set of DNA mixtures (GC-C and AC-C; GC-5mC and AC-5mC; GC-5hmC and AC-5hmC) with dogA3A and performed LC-MS/MS analysis. The results indicated that the signals for dC and 5mC were also undetectable, while the signal intensity of 5hmC was nearly unchanged following treatment with dogA3A (Fig. 3B). No significant changes were observed in the other normal nucleosides (dA, dG, and T) after treatment with either gmA3A or dogA3A (Fig. S8†). The LC-MS/MS results were consistent with those obtained from Sanger sequencing. Collectively, gmA3A demonstrated complete deamination of C and 5mC but showed no deamination activity toward 5hmC at TC and CC sites. In contrast, dogA3A also exhibited complete deamination of C and 5mC but did not deaminate 5hmC at GC and AC sites.
Quantitative evaluation of the deamination rates of 5mC and 5hmC in OMA-seq
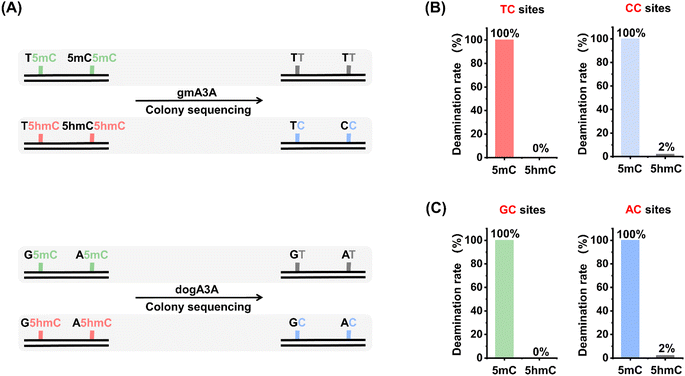
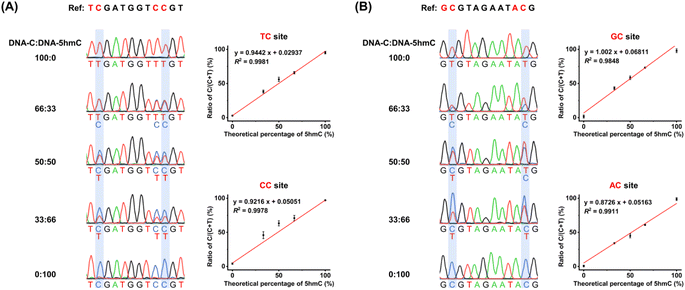
We next assessed the quantitative capability of OMA-seq. Two types of dsDNA, DNA-5mC and DNA-5hmC, were utilized as substrates, and colony sequencing was employed to evaluate the readouts of 5mC and 5hmC in OMA-seq (Fig. 4A). The colony sequencing results showed that all 5mC was fully deaminated and read as T, while 5hmC exhibited significant resistance to deamination, with a zero deamination rate at TC sites and a slightly 2% deamination rate at CC sites after treatment with gmA3A (Fig. 4B and S9†). Similarly, after treatment with dogA3A, all 5mC was fully deaminated and read as T, while 5hmC remained largely resistant to deamination, with a zero deamination rate at GC sites and a 2% deamination rate at AC sites (Fig. 4C and S10†). Since cytosine is a more suitable substrate than 5mC and both gmA3A and dogA3A can fully deaminate 5mC, cytosine can also be fully deaminated by both enzymes. Taken together, the combined use of gmA3A and dogA3A enables the distinct detection of 5hmC in all sequence contexts.Quantitative performance of OMA-seq
We quantitatively assessed the levels of 5hmC at specific sites using OMA-seq. Various mixtures of DNA-C and DNA-5hmC were prepared, with the percentage of DNA-5hmC ranging from 0% to 100%. These mixtures were analyzed using OMA-seq followed by Sanger sequencing. The measured ratio of C/(C + T) in Sanger sequencing reflects the 5hmC level in the mixture. The results indicated that the measured 5hmC levels increased proportionally to the theoretical percentages of 5hmC in the mixtures at the specified sites (Fig. 5). In conclusion, these results demonstrate that the OMA-seq method enables site-specific and quantitative detection of 5hmC in DNA.Quantitative and site-specific detection of 5hmC in genomic DNA
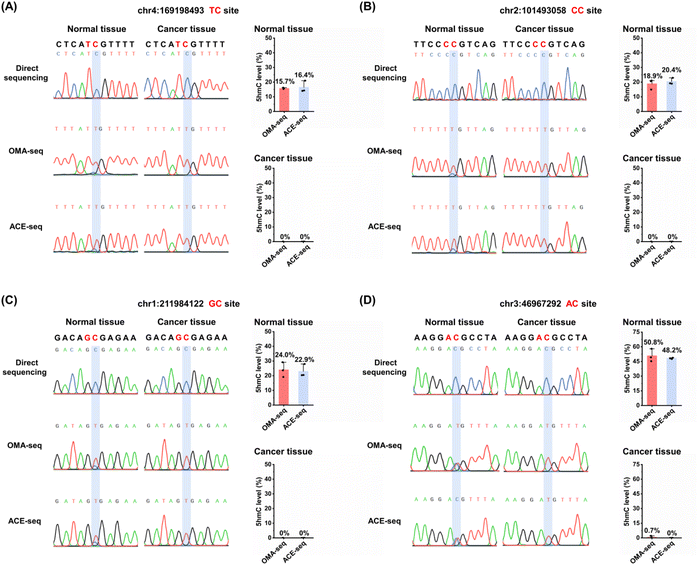
We then applied OMA-seq to detect 5hmC in specific genomic regions. Dysregulation of 5hmC has been observed in various cancers.59 Previous studies have shown that 5hmC levels are significantly reduced in cancer cell lines and primary tissues.60–62 Additionally, 5hmC signature can serve as diagnostic biomarkers for certain human cancers.63,64 In this study, we utilized the OMA-seq method for site-specific and quantitative detection of 5hmC in lung cancer tissue and its adjacent normal tissue.Since the deamination activities of gmA3A and dogA3A in the OMA-seq method can distinguish 5hmC at TC/CC sites and GC/AC sites, respectively, we selected different adjacent 5′ nucleobases of cytosine (TC, CC, GC, and AC sites) in genomic DNA for evaluation (Table S5†). 5hmC modification was reported to be present at these four sites.65 Genomic DNA was extracted and fragmented from lung cancer tissue and its adjacent normal tissue. The fragmented DNA underwent deamination treatment with gmA3A (for TC and CC sites) or dogA3A (for GC and AC sites), followed by PCR amplification. Sanger sequencing results indicated that all four sites were partially read as C and partially read as T in normal tissue (Fig. 6 and S12–S15†), suggesting the presence of 5hmC. In contrast, all four sites were completely read as T in cancer tissue (Fig. 6 and S12–S15†), indicating the absence of 5hmC at these sites. These results demonstrate that 5hmC levels are significantly reduced in cancer tissue compared to adjacent normal tissue.
We also employed the previous ACE-seq method for the quantitative analysis of 5hmC at these four sites (Fig. S11†). The quantitative results indicated that, using the OMA-seq method, the 5hmC levels in normal tissues were 15.7% at the TC site, 18.9% at the CC site, 24.0% at the GC site, and 50.8% at the AC site (Fig. 6 and S12–S15†). Similarly, the ACE-seq method detected 5hmC levels of 16.4% at the TC site, 20.4% at the CC site, 22.9% at the GC site, and 48.2% at the AC site in normal tissues. Notably, the levels of 5hmC in lung cancer tissue measured by both methods were close to zero, highlighting a significant reduction in 5hmC in cancerous tissues compared to normal tissues. The consistency of results obtained from both OMA-seq and ACE-seq underscores the reliability of these methods for quantifying 5hmC. These results are particularly important as it validates the use of OMA-seq as a robust tool for site-specific and quantitative detection of 5hmC in genomic DNA.
The introduction of new and natural enzymes for detecting 5hmC within the OMA-seq framework enhances its applicability and effectiveness. The OMA-seq method is based on a straightforward principle and features a simple analytical procedure. In OMA-seq, the deamination reaction occurs under mild conditions that prevent DNA degradation, allowing for the use of only nanogram quantities of DNA for 5hmC analysis. This technique is particularly useful when there is a limited amount of input DNA, making it ideal for site-specific and quantitative analysis of 5hmC, including applications in single-cell and cell-free DNA studies. This innovative approach, when combined with high-throughput sequencing technologies, holds great promise for enabling the quantification and genome-wide mapping of 5hmC at single-nucleotide resolution in future studies. Such advancements could significantly contribute to our understanding of epigenetic modifications in various biological contexts, including cancer research, and may lead to the identification of novel biomarkers for early diagnosis and targeted therapies.
Conclusions
We conducted a systematic characterization of the deaminase activities of six orthologous mammalian A3A proteins on C, 5mC, and 5hmC in DNA. Notably, gmA3A and dogA3A exhibited distinct deamination profiles compared to hA3A. Specifically, gmA3A completely deaminated C and 5mC at YC (Y = C/T) sites, but showed no activity towards 5hmC. In contrast, dogA3A effectively deaminated C and 5mC at RC (R = A/G) sites, with no activity on 5hmC. By combining gmA3A and dogA3A, we achieved differential deamination of C, 5mC, and 5hmC at diverse cytosine sites. Following enzyme treatment, C and 5mC were converted to T during sequencing, while 5hmC remained resistant to deamination and was still read as C. Building on these findings, we developed the OMA-seq method for quantitative and site-specific detection of 5hmC in DNA across various sequence contexts. This method enables quantitative analysis of 5hmC at individual sites in genomic DNA from lung cancer tissue and adjacent normal tissue. The OMA-seq approach is straightforward, requiring only a single-step deamination without the need for bisulfite treatment, DNA glycosylation, or chemical oxidation. This provides a valuable tool for the quantitative detection of 5hmC in DNA at single-nucleotide resolution. Furthermore, our strategy of utilizing orthologous enzymes for 5hmC detection expands the range of biological enzymes applicable in the localization analysis of modified nucleosides, offering new opportunities for epigenetic research.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Bi-Feng Yuan: writing – review & editing, investigation, supervision, conceptualization, funding acquisition. Neng-Bin Xie: writing – review & editing, investigation, supervision, conceptualization. Yu Liu: writing – review & editing, investigation, supervision, conceptualization. Xia Guo: writing – original draft, methodology, formal analysis, data curation, conceptualization. Jianyuan Wu: methodology, formal analysis, data curation, conceptualization. Tong-Tong Ji: formal analysis, data curation, conceptualization. Min Wang: methodology, data curation. Shan Zhang: methodology, data curation. Jun Xiong: data curation. Fang-Yin Gang: data curation. Wei Liu: data curation. Yao-Hua Gu: formal analysis.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The work is supported by the National Natural Science Foundation of China (22074110, 22207090), the Fundamental Research Funds for the Central Universities (No. 2042024kf0022), the Key Research and Development Project of Hubei Province (2023BCB094), and the Interdisciplinary Innovative Talents Foundation from Renmin Hospital of Wuhan University (JCRCGW-2022-008).References
- D. Schubeler, Function and information content of DNA methylation, Nature, 2015, 517, 321–326 CrossRef CAS PubMed.
- C. Luo, P. Hajkova and J. R. Ecker, Dynamic DNA methylation: In the right place at the right time, Science, 2018, 361, 1336–1340 CrossRef CAS PubMed.
- S. Kriaucionis and N. Heintz, The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain, Science, 2009, 324, 929–930 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1, Science, 2009, 324, 930–935 CrossRef CAS PubMed.
- A. Parry, S. Rulands and W. Reik, Active turnover of DNA methylation during cell fate decisions, Nat. Rev. Genet., 2021, 22, 59–66 CrossRef CAS PubMed.
- Y. Feng, Y. Q. Tian, Y. Q. Zhao, S. J. Chen and B. F. Yuan, Dynamic deformylation of 5-formylcytosine and decarboxylation of 5-carboxylcytosine during differentiation of mouse embryonic stem cells into mouse neurons, Chin. Chem. Lett., 2024, 35, 109656 CrossRef CAS.
- Y. Feng, S. J. Chen and B. F. Yuan, Recent Advances in Deciphering the Mechanisms and Biological Functions of DNA Demethylation, Chin. J. Chem., 2024, 42, 645–651 CrossRef CAS.
- Y. Feng, N. B. Xie, W. B. Tao, J. H. Ding, X. J. You, C. J. Ma, X. Zhang, C. Yi, X. Zhou, B. F. Yuan and Y. Q. Feng, Transformation of 5-Carboxylcytosine to Cytosine Through C–C Bond Cleavage in Human Cells Constitutes a Novel Pathway for DNA Demethylation, CCS Chem., 2021, 3, 994–1008 CrossRef CAS.
- O. Deniz, J. M. Frost and M. R. Branco, Regulation of transposable elements by DNA modifications, Nat. Rev. Genet., 2019, 20, 417–431 CrossRef CAS PubMed.
- E. Kriukiene, M. Tomkuviene and S. Klimasauskas, 5-Hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA, Chem. Soc. Rev., 2024, 53, 2264–2283 RSC.
- B. He, C. Zhang, X. Zhang, Y. Fan, H. Zeng, J. Liu, H. Meng, D. Bai, J. Peng, Q. Zhang, W. Tao and C. Yi, Tissue-specific 5-hydroxymethylcytosine landscape of the human genome, Nat. Commun., 2021, 12, 4249 CrossRef CAS PubMed.
- G. Ficz, M. R. Branco, S. Seisenberger, F. Santos, F. Krueger, T. A. Hore, C. J. Marques, S. Andrews and W. Reik, Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation, Nature, 2011, 473, 398–402 CrossRef CAS PubMed.
- G. D. Guler, Y. Ning, C. J. Ku, T. Phillips, E. McCarthy, C. K. Ellison, A. Bergamaschi, F. Collin, P. Lloyd, A. Scott, M. Antoine, W. Wang, K. Chau, A. Ashworth, S. R. Quake and S. Levy, Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA, Nat. Commun., 2020, 11, 5270 CrossRef CAS PubMed.
- J. Zheng and H. Wang, Highly Efficient Gel Electrophoresis for Accurate Quantification of Nucleic Acid Modifications via in-Gel Digestion with UHPLC-MS/MS, Anal. Chem., 2023, 95, 13407–13411 CrossRef CAS PubMed.
- R. Yin, J. Mo, M. Lu and H. Wang, Detection of human urinary 5-hydroxymethylcytosine by stable isotope dilution HPLC-MS/MS analysis, Anal. Chem., 2015, 87, 1846–1852 CrossRef CAS PubMed.
- W. Y. Lai, J. Z. Mo, J. F. Yin, C. Lyu and H. L. Wang, Profiling of epigenetic DNA modifications by advanced liquid chromatography-mass spectrometry technologies, TrAC, Trends Anal. Chem., 2019, 110, 173–182 CrossRef CAS.
- S. Liu and Y. Wang, Mass spectrometry for the assessment of the occurrence and biological consequences of DNA adducts, Chem. Soc. Rev., 2015, 44, 7829–7854 RSC.
- S. Liu, J. Wang, Y. Su, C. Guerrero, Y. Zeng, D. Mitra, P. J. Brooks, D. E. Fisher, H. Song and Y. Wang, Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA, Nucleic Acids Res., 2013, 41, 6421–6429 CrossRef CAS PubMed.
- Y. Yu, S. H. Zhu, F. Yuan, X. H. Zhang, Y. Y. Lu, Y. L. Zhou and X. X. Zhang, Ultrasensitive and simultaneous determination of RNA modified nucleotides by sheathless interfaced capillary electrophoresis-tandem mass spectrometry, Chem. Commun., 2019, 55, 7595–7598 RSC.
- R. Zhang, W. Lai and H. Wang, Quantification of Epigenetic DNA Modifications in the Subchromatin Structure Matrix Attachment Regions by Stable Isotope Dilution UHPLC-MS/MS Analysis, Anal. Chem., 2021, 93, 15567–15572 CrossRef CAS PubMed.
- T. Feng, Y. L. Gao, D. Hu, K. Y. Yuan, S. Y. Gu, Y. H. Gu, S. Y. Yu, J. Xiong, Y. Q. Feng, J. Wang and B. F. Yuan, Chronic sleep deprivation induces alterations in DNA and RNA modifications by liquid chromatography-mass spectrometry analysis, Chin. Chem. Lett., 2024, 35, 109259 CrossRef CAS.
- M. Y. Chen, Z. Gui, K. K. Chen, J. H. Ding, J. G. He, J. Xiong, J. L. Li, J. Wang, B. F. Yuan and Y. Q. Feng, Adolescent alcohol exposure alters DNA and RNA modifications in peripheral blood by liquid chromatography-tandem mass spectrometry analysis, Chin. Chem. Lett., 2022, 33, 2086–2090 CrossRef CAS.
- M. Berney and J. F. McGouran, Methods for detection of cytosine and thymine modifications in DNA, Nat. Rev. Chem, 2018, 2, 332–348 CrossRef CAS.
- H. Stroud, S. Feng, S. Morey Kinney, S. Pradhan and S. E. Jacobsen, 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells, Genome Biol., 2011, 12, R54 CrossRef CAS PubMed.
- C. X. Song, K. E. Szulwach, Y. Fu, Q. Dai, C. Yi, X. Li, Y. Li, C. H. Chen, W. Zhang, X. Jian, J. Wang, L. Zhang, T. J. Looney, B. Zhang, L. A. Godley, L. M. Hicks, B. T. Lahn, P. Jin and C. He, Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine, Nat. Biotechnol., 2011, 29, 68–72 CrossRef CAS PubMed.
- J. Peng, B. Xia and C. Yi, Single-base resolution analysis of DNA epigenome via high-throughput sequencing, Sci. China:Life Sci., 2016, 59, 219–226 CrossRef CAS PubMed.
- M. J. Booth, M. R. Branco, G. Ficz, D. Oxley, F. Krueger, W. Reik and S. Balasubramanian, Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution, Science, 2012, 336, 934–937 CrossRef CAS PubMed.
- M. Yu, G. C. Hon, K. E. Szulwach, C. X. Song, L. Zhang, A. Kim, X. Li, Q. Dai, Y. Shen, B. Park, J. H. Min, P. Jin, B. Ren and C. He, Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome, Cell, 2012, 149, 1368–1380 CrossRef CAS PubMed.
- K. Tanaka and A. Okamoto, Degradation of DNA by bisulfite treatment, Bioorg. Med. Chem. Lett., 2007, 17, 1912–1915 CrossRef CAS PubMed.
- C. X. Song, T. A. Clark, X. Y. Lu, A. Kislyuk, Q. Dai, S. W. Turner, C. He and J. Korlach, Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine, Nat. Methods, 2012, 9, 75–77 CrossRef CAS PubMed.
- Z. K. O'Brown, K. Boulias, J. Wang, S. Y. Wang, N. M. O'Brown, Z. Hao, H. Shibuya, P. E. Fady, Y. Shi, C. He, S. G. Megason, T. Liu and E. L. Greer, Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA, BMC Genomics, 2019, 20, 445 CrossRef PubMed.
- Y. Liu, P. Siejka-Zielinska, G. Velikova, Y. Bi, F. Yuan, M. Tomkova, C. Bai, L. Chen, B. Schuster-Bockler and C. X. Song, Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution, Nat. Biotechnol., 2019, 37, 424–429 CrossRef CAS PubMed.
- H. Xu, J. Chen, J. Cheng, L. Kong, X. Chen, M. Inoue, Y. Liu, S. Kriaucionis, M. Zhao and C. X. Song, Modular Oxidation of Cytosine Modifications and Their Application in Direct and Quantitative Sequencing of 5-Hydroxymethylcytosine, J. Am. Chem. Soc., 2023, 145, 7095–7100 CrossRef CAS PubMed.
- T. Wang, J. M. Fowler, L. Liu, C. E. Loo, M. Luo, E. K. Schutsky, K. N. Berrios, J. E. DeNizio, A. Dvorak, N. Downey, S. Montermoso, B. Y. Pingul, M. Nasrallah, W. S. Gosal, H. Wu and R. M. Kohli, Direct enzymatic sequencing of 5-methylcytosine at single-base resolution, Nat. Chem. Biol., 2023, 19, 1004–1012 CrossRef CAS PubMed.
- Y. Dai, B. F. Yuan and Y. Q. Feng, Quantification and mapping of DNA modifications, RSC Chem. Biol., 2021, 2, 1096–1114 RSC.
- J. Xiong, J. Wu, Y. Liu, Y. J. Feng and B. F. Yuan, Quantification and mapping of RNA modifications, TrAC, Trends Anal. Chem., 2024, 172, 117606 CrossRef CAS.
- J. Xiong, K. K. Chen, N. B. Xie, W. Chen, W. X. Shao, T. T. Ji, S. Y. Yu, Y. Q. Feng and B. F. Yuan, Demethylase-assisted site-specific detection of N1-methyladenosine in RNA, Chin. Chem. Lett., 2024, 35, 108953 CrossRef CAS.
- Y. H. Min, W. X. Shao, Q. S. Hu, N. B. Xie, S. Zhang, Y. Q. Feng, X. W. Xing and B. F. Yuan, Simultaneous Detection of Adenosine-to-Inosine Editing and N(6)-Methyladenosine at Identical RNA Sites through Deamination-Assisted Reverse Transcription Stalling, Anal. Chem., 2024, 96, 8730–8739 CrossRef CAS PubMed.
- X. Guo, N. B. Xie, W. Chen, T. T. Ji, J. Xiong, T. Feng, M. Wang, S. Zhang, S. Y. Gu, Y. Q. Feng and B. F. Yuan, AlkB-Facilitated Demethylation Enables Quantitative and Site-Specific Detection of Dual Methylation of Adenosine in RNA, Anal. Chem., 2024, 96, 847–855 CrossRef CAS PubMed.
- W. X. Shao, Y. H. Min, W. Chen, J. Xiong, X. Guo, N. B. Xie, S. Zhang, S. Y. Yu, C. Xie, Y. Q. Feng and B. F. Yuan, Single-Base Resolution Detection of N(6)-Methyladenosine in RNA by Adenosine Deamination Sequencing, Anal. Chem., 2023, 95, 10588–10594 CrossRef CAS PubMed.
- V. Caval, R. Suspene, J. P. Vartanian and S. Wain-Hobson, Orthologous mammalian APOBEC3A cytidine deaminases hypermutate nuclear DNA, Mol. Biol. Evol., 2014, 31, 330–340 CrossRef CAS PubMed.
- J. H. Ding, G. Li, J. Xiong, F. L. Liu, N. B. Xie, T. T. Ji, M. Wang, X. Guo, Y. Q. Feng, W. Ci and B. F. Yuan, Whole-Genome Mapping of Epigenetic Modification of 5-Formylcytosine at Single-Base Resolution by Chemical Labeling Enrichment and Deamination Sequencing, Anal. Chem., 2024, 96, 4726–4735 CrossRef CAS PubMed.
- E. K. Schutsky, C. S. Nabel, A. K. F. Davis, J. E. DeNizio and R. M. Kohli, APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA, Nucleic Acids Res., 2017, 45, 7655–7665 CrossRef CAS PubMed.
- Q. Y. Li, N. B. Xie, J. Xiong, B. F. Yuan and Y. Q. Feng, Single-Nucleotide Resolution Analysis of 5-Hydroxymethylcytosine in DNA by Enzyme-Mediated Deamination in Combination with Sequencing, Anal. Chem., 2018, 90, 14622–14628 CrossRef CAS PubMed.
- N. B. Xie, M. Wang, T. T. Ji, X. Guo, F. Y. Gang, Y. Hao, L. Zeng, Y. F. Wang, Y. Q. Feng and B. F. Yuan, Simultaneous detection of 5-methylcytosine and 5-hydroxymethylcytosine at specific genomic loci by engineered deaminase-assisted sequencing, Chem. Sci., 2024, 15, 10073–10083 RSC.
- J. Xiong, K. K. Chen, N. B. Xie, T. T. Ji, S. Y. Yu, F. Tang, C. Xie, Y. Q. Feng and B. F. Yuan, Bisulfite-Free and Single-Base Resolution Detection of Epigenetic DNA Modification of 5-Methylcytosine by Methyltransferase-Directed Labeling with APOBEC3A Deamination Sequencing, Anal. Chem., 2022, 94, 15489–15498 CrossRef CAS PubMed.
- N. B. Xie, M. Wang, T. T. Ji, X. Guo, J. H. Ding, B. F. Yuan and Y. Q. Feng, Bisulfite-free and single-nucleotide resolution sequencing of DNA epigenetic modification of 5-hydroxymethylcytosine using engineered deaminase, Chem. Sci., 2022, 13, 7046–7056 RSC.
- N. B. Xie, M. Wang, W. Chen, T. T. Ji, X. Guo, F. Y. Gang, Y. F. Wang, Y. Q. Feng, Y. Liang, W. Ci and B. F. Yuan, Whole-Genome Sequencing of 5-Hydroxymethylcytosine at Base Resolution by Bisulfite-Free Single-Step Deamination with Engineered Cytosine Deaminase, ACS Cent. Sci., 2023, 9, 2315–2325 CrossRef CAS PubMed.
- E. K. Schutsky, J. E. DeNizio, P. Hu, M. Y. Liu, C. S. Nabel, E. B. Fabyanic, Y. Hwang, F. D. Bushman, H. Wu and R. M. Kohli, Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase, Nat. Biotechnol., 2018, 36, 1083–1090 CrossRef CAS PubMed.
- F. Ito, Y. Fu, S. A. Kao, H. Yang and X. S. Chen, Family-Wide Comparative Analysis of Cytidine and Methylcytidine Deamination by Eleven Human APOBEC Proteins, J. Mol. Biol., 2017, 429, 1787–1799 CrossRef CAS PubMed.
- X. Li, V. Caval, S. Wain-Hobson and J. P. Vartanian, Elephant APOBEC3A cytidine deaminase induces massive double-stranded DNA breaks and apoptosis, Sci. Rep., 2019, 9, 728 CrossRef PubMed.
- J. Xiong, P. Wang, W. X. Shao, G. J. Li, J. H. Ding, N. B. Xie, M. Wang, Q. Y. Cheng, C. H. Xie, Y. Q. Feng, W. M. Ci and B. F. Yuan, Genome-wide mapping of N-4-methylcytosine at single-base resolution by APOBEC3A-mediated deamination sequencing, Chem. Sci., 2022, 13, 9960–9972 RSC.
- F. Tang, S. Liu, Q. Y. Li, J. Yuan, L. Li, Y. Wang, B. F. Yuan and Y. Q. Feng, Location analysis of 8-oxo-7,8-dihydroguanine in DNA by polymerase-mediated differential coding, Chem. Sci., 2019, 10, 4272–4281 RSC.
- C. J. Ma, G. Li, W. X. Shao, Y. H. Min, P. Wang, J. H. Ding, N. B. Xie, M. Wang, F. Tang, Y. Q. Feng, W. Ci, Y. Wang and B. F. Yuan, Single-Nucleotide Resolution Mapping of N(6)-Methyladenine in Genomic DNA, ACS Cent. Sci., 2023, 9, 1799–1809 CrossRef CAS PubMed.
- Y. Y. Chen, Z. Gui, D. Hu, M. Y. Chen, J. G. He, S. Y. Yu, Y. Q. Feng, J. Wang and B. F. Yuan, Adolescent alcohol exposure changes RNA modifications in adult brain by mass spectrometry-based comprehensive profiling analysis, Chin. Chem. Lett., 2024, 35, 108522 CrossRef CAS.
- X. J. You, L. Li, T. T. Ji, N. B. Xie, B. F. Yuan and Y. Q. Feng, 6-Thioguanine incorporates into RNA and induces adenosine-to-inosine editing in acute lymphoblastic leukemia cells, Chin. Chem. Lett., 2023, 34, 107181 CrossRef CAS.
- W. B. Tao, N. B. Xie, Q. Y. Cheng, Y. Q. Feng and B. F. Yuan, Sensitive determination of inosine RNA modification in single cell by chemical derivatization coupled with mass spectrometry analysis, Chin. Chem. Lett., 2023, 34, 108243 CrossRef CAS.
- X. M. Tang, T. T. Ye, X. J. You, X. M. Yin, J. H. Ding, W. X. Shao, M. Y. Chen, B. F. Yuan and Y. Q. Feng, Mass spectrometry profiling analysis enables the identification of new modifications in ribosomal RNA, Chin. Chem. Lett., 2023, 34, 107531 CrossRef CAS.
- G. Ficz and J. G. Gribben, Loss of 5-hydroxymethylcytosine in cancer: Cause or consequence?, Genomics, 2014, 104, 352–357 CrossRef CAS PubMed.
- C. E. Nestor, R. Ottaviano, J. Reddington, D. Sproul, D. Reinhardt, D. Dunican, E. Katz, J. M. Dixon, D. J. Harrison and R. R. Meehan, Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes, Genome Res., 2012, 22, 467–477 CrossRef CAS PubMed.
- C. G. Lian, Y. Xu, C. Ceol, F. Wu, A. Larson, K. Dresser, W. Xu, L. Tan, Y. Hu, Q. Zhan, C. W. Lee, D. Hu, B. Q. Lian, S. Kleffel, Y. Yang, J. Neiswender, A. J. Khorasani, R. Fang, C. Lezcano, L. M. Duncan, R. A. Scolyer, J. F. Thompson, H. Kakavand, Y. Houvras, L. I. Zon, M. C. Mihm Jr, U. B. Kaiser, T. Schatton, B. A. Woda, G. F. Murphy and Y. G. Shi, Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma, Cell, 2012, 150, 1135–1146 CrossRef CAS PubMed.
- J. Wang, J. Tang, M. Lai and H. Zhang, 5-Hydroxymethylcytosine and disease, Mutat. Res. Rev. Mutat. Res., 2014, 762C, 167–175 CrossRef PubMed.
- W. Li, X. Zhang, X. Lu, L. You, Y. Song, Z. Luo, J. Zhang, J. Nie, W. Zheng, D. Xu, Y. Wang, Y. Dong, S. Yu, J. Hong, J. Shi, H. Hao, F. Luo, L. Hua, P. Wang, X. Qian, F. Yuan, L. Wei, M. Cui, T. Zhang, Q. Liao, M. Dai, Z. Liu, G. Chen, K. Meckel, S. Adhikari, G. Jia, M. B. Bissonnette, X. Zhang, Y. Zhao, W. Zhang, C. He and J. Liu, 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers, Cell Res., 2017, 27, 1243–1257 CrossRef CAS PubMed.
- X. Tian, B. Sun, C. Chen, C. Gao, J. Zhang, X. Lu, L. Wang, X. Li, Y. Xing, R. Liu, X. Han, Z. Qi, X. Zhang, C. He, D. Han, Y. G. Yang and Q. Kan, Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer, Cell Res., 2018, 28, 597–600 CrossRef CAS PubMed.
- X. Li, Y. Liu, T. Salz, K. D. Hansen and A. Feinberg, Whole-genome analysis of the methylome and hydroxymethylome in normal and malignant lung and liver, Genome Res., 2016, 26, 1730–1741 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: In vitro expression and purification of orthologous mammalian A3A proteins; colony sequencing; enzymatic digestion of DNA; LC-MS/MS analysis; Tables S1–S5; Fig. S1–S15. See DOI: https://doi.org/10.1039/d4sc08660k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |