Open Access Article
Open Access ArticleCyanobacterial green chemistry: a blue-green approach for a sustainable environment, energy, and chemical production
Priyul
Pandey†
,
Deepa
Pandey†
,
Anjali
Gupta
,
Rinkesh
Gupta
,
Sapna
Tiwari
and
Shailendra Pratap
Singh
 *
*
Department of Botany, Institute of Science, Banaras Hindu University, Varanasi-221005, UP, India. E-mail: spsingh@bhu.ac.in
First published on 14th January 2025
Abstract
Increased human activity due to the ever-increasing global population has necessitated the urgent need for a sustainable environment, food, and energy. Cyanobacteria, classically known as blue-green algae, are oxygen-producing photosynthetic organisms that are emerging as an option to achieve sustainable development goals. These Gram-negative prokaryotes can efficiently sequester atmospheric CO2 due to an efficient carbon concentrating mechanism and divert it to the production of energy-rich compounds, i.e., biofuel, and other valuable chemicals, using their flexible metabolic chassis. Additionally, cyanobacteria also minimize the emission of methane, which is another greenhouse gas, by providing oxygen to methane-oxidizing bacteria. In recent years, several genetically engineered strains of cyanobacteria have been developed that can produce biofuels and several other valuable chemicals. Strains have also been engineered for bioplastic production and bioremediation purposes. These organisms have gained attention as biofertilizers and can increase the quality and fertility of soil. Thus, cyanobacteria are promising CO2 sinks that can contribute to global efforts in carbon capture and storage initiatives while producing bioenergy, cosmetics, pharmaceuticals, and several other valuable chemicals. Therefore, these blue-green cells can be used for green chemistry while minimizing the atmospheric CO2 concentration. In this review, we present various applications of cyanobacterial biomass to achieve sustainable development goals. We also discuss challenges associated with the wide application of cyanobacteria and the future direction to make full use of these robust organisms to fulfill our future demands in an environment-friendly manner.
Sustainability spotlightThe sustainability of the environment, energy, and food is a major challenge associated with the continuous increase in global population. The greenhouse gas emissions due to fossil fuel burning and other industrial processes require a carbon-neutral society. The overexploitation of chemical fertilizers and other industrial contaminants is compromising the health of air, water, and soil. Also, the continuous piling of plastic waste, resistant to natural breakdown, is another environmental challenge. To overcome these challenges, greener ways of CO2 sequestration, fertilizers, chemicals, and energy production are required without causing any negative impact on air, water, and soil. Cyanobacteria and microalgae are emerging biosystems that have the potential to fulfill future demands of sustainable food, energy, and the environment. In this work, we have discussed different features of cyanobacteria that can be used to sequester CO2, and sequestered CO2 can be diverted through their versatile metabolic chassis using synthetic and molecular biology tools to produce food, energy, valuable chemicals, bioplastics, and water treatment systems. This work emphasizes the importance of the following UN sustainable development goals: zero hunger (SDG 2), good health and wellbeing (SDG 3), clean water and sanitation (SDG 6), affordable and clean energy (SDG 7), industry, innovation, and infrastructure (SDG 9), climate action (SDG 13), and life on land (SDG 15). |
1 Introduction
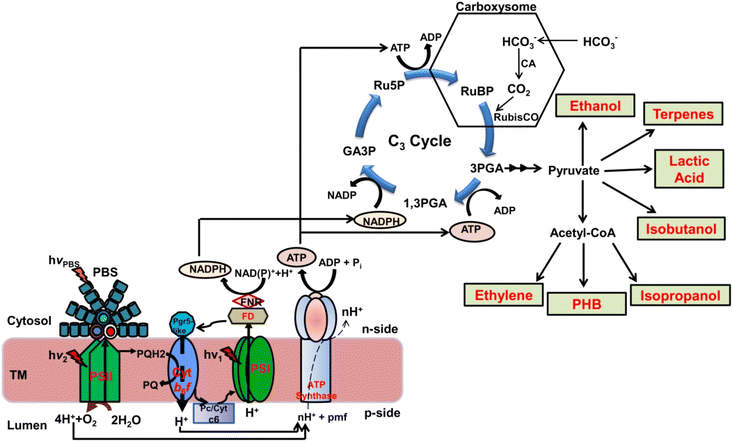
The global population is continuously increasing and recently India has become the most populated country on the planet Earth.1 The ever-increasing population has both advantages and disadvantages. The advantage is to become a vast and growing consumer market to attract investment, increased workforce, and domestic production. However, along with these advantages, there are several disadvantages such as unemployment, poverty, scarcity of food, environmental degradation, overexploitation of natural resources, global warming, and environmental fluctuations, i.e., climate change.2 Climate change presents a multifaceted threat to global ecologies, societies, and economies and affects various environmental aspects such as extreme weather events, ice sheet melting, and sea level rise.3–5 In response to the abovementioned environmental crisis, the Paris Agreement 2015 aimed to combat climate change and accelerate investments and actions to achieve a low-carbon sustainable future. Therefore, the rise in global temperature due to carbon dioxide (CO2) emissions and other anthropogenic gases underscores the urgent need for action to combat the negative effects of climate change.6In order to avoid damages from global warming and climate change, it is urgently required to limit greenhouse gas (GHG) emissions, particularly CO2, from various sources, in addition to sequestering it into biomass.7 In the last two decades, cyanobacteria, classically known as blue-green algae due to their characteristic blue-green color, have emerged as promising organisms for the sustainability of the environment, agriculture, and energy.7–9 Cyanobacteria play a crucial role in mitigating GHG emissions, mainly through CO2 sequestration. As oxygenic photosynthetic Gram-negative prokaryotes, cyanobacteria utilize sunlight to convert CO2 into organic compounds (Fig. 1), thereby controlling its concentration in the atmosphere.10,11 Thus, cyanobacteria can act as a CO2 sink due to the presence of a very effective carbon-concentrating mechanism (Fig. 1). These organisms accumulate inorganic carbon in the form of bicarbonate inside the cells in a proteinaceous microcompartment called a carboxysome.11–13
The special feature of efficiently sequestering atmospheric CO2 makes cyanobacteria valuable allies in combating climate change and promoting environmental sustainability. In addition to CO2, cyanobacteria can also mitigate methane production, which is another GHG, via modulating microbial methane metabolism. Cyanobacteria promote the growth of methane oxidizing microorganisms (MOB) and utilize their metabolites. The physicochemical properties of soil are improved by cyanobacteria that can further promote methane mitigation by MOB.14 In the context of climate change mitigation, cyanobacterial role as a potent CO2 sink provides a valuable tool for reducing atmospheric carbon levels. The efficient photosynthetic conversion of CO2 coupled with high biomass production by cyanobacteria makes these organisms a key player in biological carbon sequestration efforts.7,11,15
In the last few decades, efforts have been made to enhance the usage of cyanobacteria by using genetic manipulation and synthetic and metabolic engineering approaches.16 Through genetic engineering, cyanobacteria produce various kinds of chemicals, including alcohols, hydrocarbons, proteins, carbohydrates, carboxylic acids, isoprenes, toxins, antioxidants, pigments, vitamins, and methane. These chemicals have different applications in the biofuel, food, pharmaceutical, and cosmetic industries. Cyanobacteria are also useful in bioremediation and industrial biotechnology.16,17 Moreover, the use of cyanobacteria as biofertilizers offers a sustainable approach to enhancing crop yields by improving soil fertility and decreasing methane emissions in agricultural ecosystems such as rice fields. By leveraging the unique nitrogen-fixing abilities of cyanobacteria, farmers can enhance soil health, reduce the need for chemical fertilizers, and therefore, can contribute to environment-friendly agricultural practices.18,19 These approaches not only benefit food security but also help in mitigating GHG emissions associated with conventional farming practices.
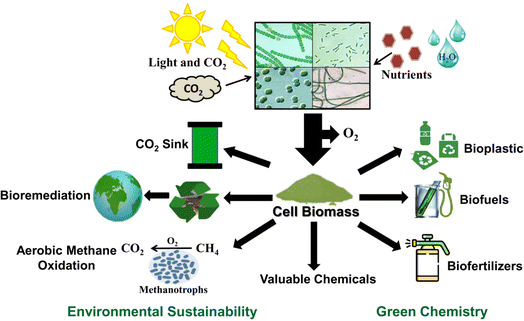
Cyanobacteria, with their unique capabilities and diverse applications particularly in environmental sustainability along with industrial biotechnology, hold significant promise for addressing critical global challenges such as climate change, food security, GHG emissions, and bioenergy production (Fig. 2). Thus, this review highlights the multifaceted role of cyanobacteria in achieving sustainability of the environment, energy, and chemical production while minimizing the levels of CO2 and methane and fixing atmospheric dinitrogen.
2 Cyanobacterial relevance in reducing greenhouse gas emissions
Cyanobacteria play a significant role in reducing GHG emissions through their unique metabolic processes. These photosynthetic microorganisms play a crucial role in carbon sequestration by absorbing atmospheric CO2 during photosynthesis, converting it into organic molecules and biomass (Fig. 1 and 2), and releasing oxygen as a byproduct. Additionally, several cyanobacteria are capable of nitrogen fixation, and therefore, these organisms can enhance soil fertility and support plant growth while contributing to global carbon fixation.20 Also, cyanobacteria and their genetically engineered strains have been explored for their potential in biofuel production to offer a renewable and carbon-neutral alternative to fossil fuels.7,8 By harnessing their capabilities in biofertilizers, methane reduction, carbon sequestration and fixation, and sustainable bioenergy production, cyanobacteria offer promising avenues for mitigating GHG emissions and addressing problems associated with global climate change (Fig. 2).2.1 Cyanobacterial oxygenation boosts methanotroph-driven methane oxidation in wetland rice cultivars
In present days, the discussion and research on methane production, release, and usage have gained significant attention due to its impact on global warming and atmospheric chemistry.21 Agricultural fields such as wetland rice fields, rivers, estuaries, oceans, wildfires, vegetation, and animals are identified as major natural sources of methane emission. In addition to natural sources, the majority of anthropogenic activities such as biomass burning and landfilling, animal rearing, and consumption of fossil fuels contribute to global methane emission.22 Also, methane emissions in the atmosphere can increase as a result of population growth, waste production, and increased demand for food and fossil fuels. Nonetheless, methanogenesis in anaerobic flooded rice fields makes these agroecosystems one of the most important contributors to methane production.23Agriculture is essential for both financial and food security, but it needs to be environment friendly. Therefore, to feed the increasing global population, the production of rice grains in the future will require the use of environment-friendly fertilizers and genetically engineered rice cultivars that can trap more carbon in the grains to minimize the emission of methane.24 It is imperative to increase agricultural productivity without sacrificing the sustainability of the environment, and therefore, even a simple strategy can make a significant difference. According to Cuellar-Bermudez (2019),25 cyanobacteria hold great promise for addressing the problem of global warming which is caused by GHG released due to natural and human activities. In addition to lowering the required quantity of nitrogen fertilizers, cyanobacteria can increase rice production with a simultaneous reduction in the amount of methane generated during rice cultivation.26
Thus, in the agricultural ecosystem, the applications of cyanobacteria can increase the production of grains, maintain the fertility and texture of the soil, and decrease methane yield. The methane produced by methanogenic bacteria in rice fields is utilized by methanotrophs which are obligate aerobic autotrophic microorganisms.27 Methanotrophs convert methane into CO2 to obtain energy from its oxidation.28 The CO2 produced during the oxidation of methane can be consumed by cyanobacteria. Thus, methanotrophs play an important role in eliminating methane produced during rice cultivation or from any other sources. However, the oxidation of methane in water-logged habitats is limited by the availability of oxygen.29 Therefore, inoculation of cyanobacteria and/or algae in water-logged conditions, particularly in rice fields, can overcome the limitation of oxygen and promote the oxidation of methane by methanotrophs. In addition to promoting the oxidation of methane, the use of Anabaena sp., Nostoc sp., Calothrix sp., and Tolypothrix sp. as cyanobacteria-based biofertilizers enriches the soil quality and fertility by secreting several chemicals and fixing atmospheric nitrogen.7,30 However, excessive use of chemical fertilizers can limit the growth of microorganisms and, therefore, can negatively impact soil quality as well as consumption of methane by methanotrophs.
As a proof-of-concept, it has been shown that methane emission can be decreased by the use of cyanobacterial/algal biofertilizers due to the availability of oxygen produced during photosynthesis by these organisms to increase the oxidation of methane.30 Thus, the integrated approach of using cyanobacterium-based biofertilizers not only improves the quality and fertility of the soil but also decreases the net production of methane from the agricultural field to attain sustainable agricultural practices.
2.2 Harnessing cyanobacteria as CO2 sinks: an approach to climate sustainability
The primary cause of increasing CO2 levels in the atmosphere is the burning of fossil fuels such as petroleum, coal, and natural gas which ultimately leads to global warming and climate change.7,31,32 Human activities, particularly industrialization and excessive fossil fuel burning mainly due to transportation, have significantly increased GHG emissions.7 To address the problems associated with global warming and climate change, various physical and biological methods have been employed to mitigate CO2 levels.Physical means of CO2 sequestration have many disadvantages: having high costs associated with capturing, transporting, and storing CO2. However, biological sequestration of CO2 using cyanobacteria has been proposed to be a highly effective approach for decreasing the level of atmospheric CO2.7,33,34 The CO2 sequestered by cyanobacteria can be diverted for the production of valuable green chemicals, nitrogen or protein-rich biomass, and biofuels using flexible metabolic chassis of wild-type or genetically engineered strains (Fig. 1).7,8,15 Cyanobacteria are selected primarily due to their efficient CO2 sequestration ability and photosynthetic conversion of CO2 to sugar, various industrially important bioactive molecules, and biomass production.8,11,15,35
Cyanobacteria are found in diverse habitats, including marine and freshwater ecosystems, and possess a carbon-concentrating mechanism (CCM) that enables them to accumulate a ∼1000 times higher concentration of CO2 than the ambient level.11,12,35,36 The absorbed CO2 is accumulated around the Rubisco enzyme in a proteinaceous microcompartment called a carboxysome where its further reduction takes place to lock the sequestered CO2 into simple sugar.15,37 The cyanobacterial CCM involves transporters for inorganic forms of carbon (Ci) such as CO2 and HCO3− along with carboxysomes that encapsulate Rubisco and carbonic anhydrase (CA) enzymes.11,12,38 It is believed that the cyanobacterial CCM evolved to minimize photorespiration that came into effect due to changes in the absolute and relative levels of CO2 and oxygen in the environment after the evolution of oxygenic photosynthesis.11,12
The cyanobacterial genome codes for Ci transporters as single-gene products (e.g., BicA and SbtA) and some of the operons coding for transport machinery (e.g., BCT1, NDH-13, and NDH-14). Cyanobacteria have two CO2 absorption systems located in the thylakoid membrane and three transporters in the plasma membrane.11–13 Together, these five distinct inorganic carbon transporters make the CO2 acquisition system of cyanobacteria. However, all cyanobacteria may not have all five transporters that differ from each other in terms of their unique net affinity and uptake flux capacity for CO2 sequestration inside the cell.12,13 In order to mitigate the impact of growing CO2 concentrations in the atmosphere, the potential of cyanobacteria to sequester CO2 due to the presence of an efficient CCM is getting recognition. Cyanobacteria play a significant role in the overall photosynthetic conversion of solar energy and CO2 assimilation which is almost ten to fifty times faster than in terrestrial plants.39 Therefore, using the potential of these biological systems is one of the most promising ways to lower the atmospheric CO2 level to mitigate the negative impacts of global warming.7 Currently, trees are a potential carbon capture solution in cities; however, their usage to capture CO2 may be restricted due to their density, rate of growth, and availability of space.
Also, the high levels of atmospheric CO2 together with other contaminants can promote stomatal closure to affect the uptake of CO2 by plants.39 The availability of green space is continuously decreasing in urban areas due to increasing population density, buildings, and other constructions. Therefore, an alternative approach must be taken to ensure that cities have a zero-carbon balance. The method of utilizing cyanobacteria/algae to capture CO2 and other toxins from air, soil, and water into their biomass is called bio-capture or phyco-capture of CO2.7,40,41 As an alternative to large areas occupied by trees, the growth of cyanobacteria in photobioreactors has gained attention as an alternative approach to capturing carbon.42
It is estimated that almost half of the oxygen available in the atmosphere is produced by cyanobacteria and microalgae which is released during the conversion of CO2 and solar radiation into chemical energy in the form of sugars.15,36 According to Farrelly et al. 2013,43 these organisms can absorb up to 2.35 gigatons (Gt) of CO2 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 km3 culture area. It is also suggested that carbon fixation by microalgae could be a significant proportion of the remaining unidentified carbon sink.44 Thus, cyanobacteria together with other microalgae have the potential for CO2 sequestration to tackle side-effects of global warming due to CO2 emission.
000 km3 culture area. It is also suggested that carbon fixation by microalgae could be a significant proportion of the remaining unidentified carbon sink.44 Thus, cyanobacteria together with other microalgae have the potential for CO2 sequestration to tackle side-effects of global warming due to CO2 emission.
3 Cyanobacteria can promote sustainable agricultural practices
The Green Revolution entered the global economy in 1965 and opened up new avenues for the agriculture sector which is now regarded as the backbone of the economy and provides several opportunities to a large portion of the population.45 While excessive use of chemical fertilizers has a positive effect on crop productivity, it could negatively impact the structure and physiochemical properties of the soil as well as the biotic and abiotic components of an ecosystem. These chemicals affect beneficial microbes along with nematodes and insects that are known to improve soil properties.45,46 However, in the current scenario of continuous increase in global population, there is a need to further increase global food production without compromising the environment. Therefore, microbe-based biofertilizers present an excellent alternative to chemical-based fertilizers for sustainable agricultural practices.19Microbe-based fertilizers can reduce the usage of chemical fertilizers and increase crop production by improving the quality and fertility of soil. Among microbe-based fertilizers, several cyanobacteria are considered potent biofertilizers because of their ability to fix atmospheric nitrogen, secrete secondary metabolites, and improve soil fertility and structure.19,47 Cyanobacterial biomass enhances the physico-chemical properties of deteriorated soil and increases soil water-holding capacity and mineral nutrient status.48 By naturally fixing atmospheric nitrogen, solubilizing phosphates, and producing chemicals that promote plant growth, biofertilizers supplement soil with additional nutrients. Biofertilizers are substances that hold microbial inoculants, artificially multiplied cultures of certain soil microbes that colonize the rhizosphere. However, by increasing the supplement or availability of primary micro- and macronutrients and growth-stimulating agents on the target crop, these microbial inoculants enhance soil fertility and crop productivity.49
There are approximately 1011 microbial cells per gram and more than 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 prokaryotic species found in the rhizosphere which is a narrow zone of soil that surrounds plant roots and generally contributes to plant productivity.49,50 In a natural agro-ecosystem, the interaction between bacteria and plants in the rhizosphere influences crop health by offering many services to crop plants such as decomposing organic matter, acquiring nutrients, absorbing water, recycling nutrients, and controlling weeds.19,51 In addition to cyanobacteria, plant growth-promoting rhizobacteria (PGPR), phosphorus-solubilizers, sulfur-oxidizers, mycorrhiza, bacteria that can suppress diseases, endophytes that can withstand stress, and decomposers of organic matter are examples of other bioinoculants that function as biofertilizers.52 Thus, cyanobacterial application in agricultural fields can promote sustainable agricultural practices by increasing soil quality and fertility without using chemical fertilizers.
000 prokaryotic species found in the rhizosphere which is a narrow zone of soil that surrounds plant roots and generally contributes to plant productivity.49,50 In a natural agro-ecosystem, the interaction between bacteria and plants in the rhizosphere influences crop health by offering many services to crop plants such as decomposing organic matter, acquiring nutrients, absorbing water, recycling nutrients, and controlling weeds.19,51 In addition to cyanobacteria, plant growth-promoting rhizobacteria (PGPR), phosphorus-solubilizers, sulfur-oxidizers, mycorrhiza, bacteria that can suppress diseases, endophytes that can withstand stress, and decomposers of organic matter are examples of other bioinoculants that function as biofertilizers.52 Thus, cyanobacterial application in agricultural fields can promote sustainable agricultural practices by increasing soil quality and fertility without using chemical fertilizers.
4 Cyanobacterial CO2 capture: a green pathway to renewable energy rich chemical production
The whole world is looking towards options for renewable energy production to combat global climate change, limited availability of conventional energy resources, and their impact on GHG emissions. The recent G20 conference held in New Delhi witnessed the creation of the India-led Global Biofuels Alliance (GBA) in order to encourage the use of environment-friendly biofuels. The aim of the GBA is to form a coalition between governments, international organizations, and business partners to promote the production and consumption of biofuels. It is estimated that global biofuel production needs to be increased three times to put the world energy system back on track with net zero carbon emission by 2050.7,53The rise in biofuel demand is driven by several factors, including increasing oil prices, the urgent need to reduce GHG emissions from fossil fuel burning, the quest for energy independence among nations, and the opportunity for farmers to make a profit by providing feedstock and agricultural waste for biofuel production.54 Additionally, the production and utilization of biofuels are recognized as a carbon-neutral approach as they are produced from biomass that has sequestered a substantial amount of CO2 from the atmosphere.7,55 Globally, there is a growing trend in the production of biofuels from diverse bioresources through the application of innovative technology and biological processes. The utilization of agricultural crop waste and biomass to produce biofuels has been anticipated to lessen the negative impact of fossil fuel burning on the environment and the disposal of agricultural waste.7,56
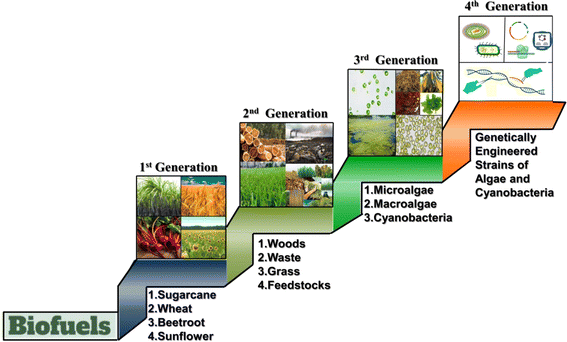
Therefore, the production of biofuels from plants and microbial biomass has been the subject of investigation in the last few decades.57,58 However, the production of biofuel from agricultural crops or waste can result in competition for fertile land to produce food and energy. To overcome this competition, additional research has been conducted on cutting-edge technology for the production of biofuels from alternative feedstock. Based on the type of feedstock used for the production of biofuels, there are four categories of biofuels (Fig. 3), i.e., first (1G), second (2G), third (3G), and fourth (4G) generations of biofuels.8,59 A range of both gaseous and liquid biofuels such as biodiesel, ethanol, methanol, and methane are currently being produced from different biomass feedstocks.60–62 Also, bioenergy produced from biomass is anticipated to contribute around 10–50% of the world's energy consumption by 2050.7 Therefore, biomass-based energy production together with other sources of renewable energy such as solar and wind energy will play a significant role in fulfilling future global energy demand in a sustainable and environment-friendly manner.
The 1G biofuel is produced from edible food crops such as sugarcane, potato, oilseed, corn, barley, wheat, sunflower, and soybean.7,8 Ethanol was the first energy-rich chemical produced from raw corn and sugarcane using fungal mycelia as a source for fermentation of sugars.63,64 Fungi, e.g., Rhizopus sp. and Saccharomyces cerevisiae, can produce ethanol by fermentation of raw corn flour, and consequently, bioethanol has been produced on a large-scale from starch through enzymatic hydrolysis.65,66 The 2G biofuel is produced from cellulose and different organic waste materials such as wood, straw, and switchgrass and non-food plants such as Jatropha seeds.7,8,67 3G biofuels are produced from eukaryotic algae and cyanobacterium feedstock, while the 4G biofuel production involves the use of genetically engineered strains of algae and cyanobacteria.7,8 Genetically engineered strains having higher biomass production than their native strains or any other phenotypic alteration that gives them a competitive advantage over other strains are better suited for bioenergy production.
The quest for new energy alternatives has been further fueled by the urgent need to decrease the emission of CO2 from fossil fuel burning. Therefore, genetically engineered strains of cyanobacteria and eukaryotic algae having higher CO2-sequestration efficiency can give an alternate option to energy sources while sequestering atmospheric CO2.11,68 The algal or cyanobacterial biomass contains a large amount of carbohydrates, lipids, proteins, and several other constituents that can be used to produce different types of biofuels using physical and biological methods.7 However, compared to algae and other photosynthetic organisms, cyanobacteria offer several advantages in biofuel production due to high growth rates leading to increased feedstock production, lower water, fertile land demand, simple and cost-effective nutritional requirements, and the ease of genetic manipulation through available trusted and tested genetic engineering tools.7,8,62
Biofuels derived from cyanobacteria offer a blend of alkanes, fatty acids, and fatty alcohols that closely resemble fossil fuels. This similarity of energy-rich molecules suggests that these biofuels could serve as excellent substitutes for transportation fuels without requiring significant alterations to vehicle engines.58 Over the past few decades, several attempts have been made to use photosynthetic organisms as living tools to convert solar energy into renewable energy. The production of ethanol by engineered Synechococcus elongatus PCC 7942, incorporating pyruvate decarboxylase and alcohol dehydrogenase II encoding genes from Zymomonas mobilis into its genome, marked a significant milestone in energy production using cyanobacteria.69,70 Algenol Biotech, which is an industrial biotechnology company, established in 2009 in Fort Myers, FL, USA, uses genetically engineered cyanobacterial strains to produce bioethanol.
The genetic manipulation of basic cyanobacterial metabolic chassis has produced a number of compounds valuable for the energy sector (Fig. 1 and 4). The major advancement was made by diverting the carbon flux towards the production of ethanol in Synechococcus elongatus PCC 7942.69,71 However, ethanol is not the greatest substitute for gasoline due to its lower energy content and ability to absorb water.72 Therefore, the focus has been shifted to developing energy-rich molecules with longer carbon chains. For example, isobutyraldehyde, which is a crucial ingredient for producing fuels from petroleum, has been produced in an efficient manner by modifying the valine biosynthetic pathway in S. elongatus PCC 7942.73 Despite these advantages, there are environmental concerns associated with the use of genetically modified (GM) cyanobacteria or any other microalgal strain for biofuel production. Potential issues include horizontal gene transfer and competition between GM strains and other microorganisms which could impact the natural balance of an ecosystem. However, large-scale cultivation of GM strains using closed photobioreactors can overcome the abovementioned challenges, but the economic viability of large-scale cultivation using expensive closed photobioreactors only for biofuel production still needs to be achieved.7,8 Therefore, an integrated approach for the production of bioenergy and value-added molecules has been proposed to achieve economic viability.8 Also, the policy related to the use of genetically engineered strains of cyanobacteria and algae is still not clear at the global level.
5 Cyanobacteria in wastewater treatment: an eco-friendly approach for bioremediation
The global concern for the environment and human well-being has increased in the last few decades due to the addition of different pollutants to waterways.74 Cyanobacteria exhibit versatile capabilities in addressing environmental issues, particularly in wastewater management and soil remediation. These microorganisms contribute to removing excess levels of nitrogen and heavy metals from water systems and, therefore, have the ability to combat eutrophication. Cyanobacteria can metabolize complex compounds such as hydrocarbons and pesticides, and therefore, these organisms can be useful in promoting soil and water restoration.75–77Domestic wastewater containing paper, human excrement, and synthetic detergent residues intermixes with industrial effluents containing a variety of organic and inorganic pollutants.78 Pesticides following their application permeate natural basins through soil erosion or atmospheric precipitation. Organic matter and suspended solids in wastewater impair light penetration and, therefore, hamper photosynthesis, promote sediment buildup, disrupt water self-purification processes, and diminish dissolved oxygen levels, whereas surface-active agents such as fatty acids and oils lower the oxygen content of water by producing a film layer.78
To address these issues, attention has been paid to biological wastewater treatment using cyanobacteria.79 Cyanobacteria exhibit the exceptional capacity to decompose oil constituents and complex organics and sequester ions of zinc, cobalt, and copper metal.77,80 Therefore, cyanobacteria exhibit promising solutions for treating secondary effluents derived from urban, agricultural, or industrial sectors. These microorganisms serve as powerful agents for the restoration and cleaning of diverse environmental settings as their distinctive metabolic characteristics and adaptive mechanisms allow them to effectively handle various pollutants.19,28 In aquatic ecosystems, cyanobacteria excel in purifying contaminated water bodies by absorbing heavy metals such as mercury, lead, and cadmium, and therefore, these organisms can effectively minimize the hazardous consequences of these toxic substances in aquatic ecosystems.77,81,82
However, optimization of cyanobacterial strains for specific pollutants, ensuring ecological safety, and scaling up bioremediation processes for large-scale applications still remain big challenges. To fully harness the potential of cyanobacteria, collaborative efforts involving genetic engineering and an enhanced understanding of their physiology are essential.83 This will permit the use of cyanobacteria in the ongoing fight against environmental degradation and conservation efforts. Cyanobacteria such as Oscillatoria, Phormidium, Aphanocapsa, and Westiellopsis have demonstrated their capability to eliminate nitrogen and phosphate ions from wastewater.84 Additionally, Spirulina strains possess diverse functional groups such as carboxyl, hydroxyl, and sulfate, which facilitate metal binding. Thus, the presence of these functional groups makes Spirulina strains effective for controlling pollutants such as zinc and nickel.84
Also, research indicates that certain cyanobacteria such as Synechocystis sp., Westiellopsis prolifica, Nostoc hatei, and Anabaena sphaerica can break down organophosphorus or organochlorine insecticides in aquatic environments.84 However, widespread implementation of cyanobacteria in wastewater treatment is limited due to the efficient removal of biomass following treatment. Immobilization techniques using agarose, carrageenan, chitosan, alginate, and polyurethane foam can enhance the efficiency of nutrient or metal uptake with efficient removal of biomass from treated water.85 In natural ecosystems, several cyanobacterial species such as those belonging to genera such as Oscillatoria, Synechocystis, and Pleurocapsa form symbiotic associations with both aerobic and anaerobic microorganisms.84 Notably, cyanobacterial communities contribute to the breakdown of hydrocarbon pollutants found in crude oil spillover.85 While cyanobacteria do not directly decompose hydrocarbons, these organisms facilitate the process by supplying essential resources such as oxygen and nutrients to oil-degrading bacteria in a consortium.85
Therefore, to enhance the effective decomposition of hydrocarbons, an engineered consortium consisting of multiple cyanobacterial species, including Phormidium, Oscillatoria, and Chroococcus and the oil-degrading bacterium Burkholderia cepacia has been successfully developed and proven its efficiency for decomposing petroleum compounds.86
6 Polyethylene biodegradation: a sustainable approach to reducing plastic pollution
Polyethylene is a widely used material due to its affordability, durability, and versatility in various applications such as packaging, textile transportation, and manufacturing. However, its wide application is a major environmental challenge due to its improper disposal, especially in marine and urban environments.87 Furthermore, its resistance to natural degradation is yet another environmental challenge. Due to the challenges encountered in the recycling of plastic waste, bioremediation gives an alternative approach to the breakdown of polyethylene by microorganisms.88 In this process, microalgae and cyanobacteria can adhere to the surface of submerged polyethylene and promote biodegradation by secreting substances that aid in the colonization of microorganisms.88 Some microorganisms can even directly use polyethylene as the sole carbon source.89 While bacteria and fungi have been extensively studied for polyethylene biodegradation, research on potential of cyanobacteria and other algae remains limited.90Notably, cyanobacteria and algae such as Anabaena spiroides, Scenedesmus dimorphus, and Navicula pupula have shown potential in degrading polyethylene.90 According to Sarmah and Rout 2018,88 two cyanobacterial species such as Phormidium lucidum and Oscillatoria subbrevis found on algal-covered polyethylene surfaces in sewage water are capable of utilizing polyethylene as a carbon source without any additional treatment. Thus, cyanobacteria together with microalgae offer a promising avenue for developing eco-friendly polyethylene degradation technology that is efficient, safe, and easy to implement. However, further intensive work is required to explore the possibility of using cyanobacteria and microalgae for the biodegradation of polyethylene.
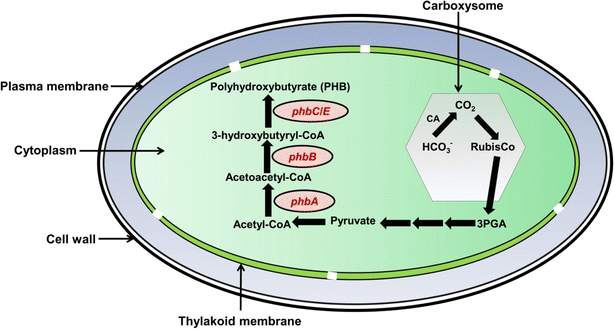
7 Polyhydroxybutyrate (bioplastic) production and biodegradation
The degradation of polyethylene is extremely slow, and therefore, biodegradable polymers can be an alternative to a polyethylene-free environment. Polyhydroxybutyrate (PHB) is a biodegradable polymer that has gained significant attention due to its potential application as a sustainable alternative to conventional plastics in various industries.91 Cyanobacteria possess metabolic pathways that enable them to synthesize PHB as an intracellular carbon storage compound.92 PHB biosynthesis occurs via the condensation of acetyl-CoA molecules into D-3-hydroxybutyryl-CoA followed by polymerization and accumulation of PHB granules within cyanobacterial cells.93 Understanding the molecular mechanism and regulatory factors governing PHB biosynthesis in cyanobacteria is crucial for optimizing PHB production. Different cyanobacterial species such as Spirulina platensis, Gloeothece sp., Oscillatoria limosa, Synechococcus sp., and Synechocystis sp. accumulate considerable amounts of PHB.91PHB is produced by three enzymatic reactions that start from acetyl coenzyme A (Fig. 1 and 4). Two acetyl-CoA molecules are condensed and converted to one acetoacetyl-CoA molecule by a 3-ketothiolase enzyme encoded by the phbA gene.91 Furthermore, acetoacetyl-CoA is converted to D-3-hydroxybutyryl-CoA by NADPH-dependent acetoacetyl-CoA reductase which is encoded by the phbB gene. The final enzyme PHA synthase, encoded by phaC/phaE, forms PHB and also catalyzes the ester bond-forming process between the D-3-hydroxybutyryl moiety and an already-existing PHB molecule.91 The enzymatic biodegradation of PHB by microorganisms such as algae, fungi, and bacteria involves a set of reactions that starts with the breaking of primary bonds of the polymer. This results in a change in the chemical structure of PHB and reduces its molar mass.92 The degradation of PHB begins with the action of PHA depolymerase, encoded by the phaZ gene, which acts on the PHB molecule and releases intracellular D-3-hydroxybutyrate. Furthermore, this molecule is oxidized by the action of 3-hydroxybutyrate dehydrogenase to produce acetoacetate which is further esterified to acetoacetyl-CoA.92 Acetoacetyl-CoA is hydrolyzed by the phaB gene product to form acetyl-CoA which enters the tricarboxylic acid (TCA) cycle and ultimately degrades to CO2 and H2O. The degradation of PHB produces CO2 and H2O under aerobic conditions whereas CO2 and CH4 are produced under anaerobic conditions.94
However, despite significant progress, several challenges remain to be addressed for the widespread commercialization of cyanobacterium-derived PHB.95 These challenges include optimizing the rate of PHB production, improving the robustness and stability of cyanobacterial strains under dynamic environmental conditions, and reducing its production cost. Additionally, the development of scalable cultivation systems and downstream processing methods is essential for the industrial-scale production of PHB.96 Therefore, future research efforts should focus on addressing the abovementioned challenges and exploring novel strategies for enhancing PHB production using cyanobacteria. Also, attention should be paid to screening new cyanobacterial isolates capable of producing PHB.
8 Cyanobacteria are biofactories for valuable chemical production (green chemistry)
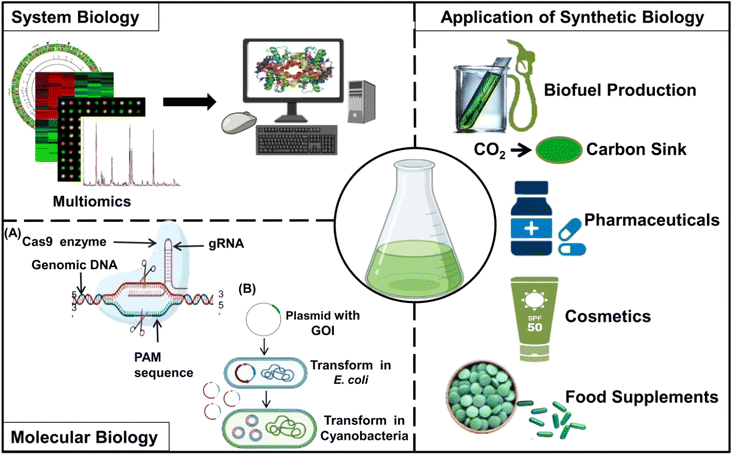
The potential of genetically engineered cyanobacterial strains to directly convert CO2 into target compounds provides an environment-friendly and sustainable approach for green chemistry to produce various valuable chemicals (Fig. 1). Cyanobacteria offer an immense potential for genetic/metabolic engineering to produce a range of chemicals due to their unique features such as smaller genomes and genetic/metabolic diversity, rapid growth rates, and efficient mechanisms for CO2 absorption.8,97 Cyanobacteria are known to naturally produce a variety of enzymes and chemicals required for the functioning of their basic metabolic pathways. However, fine-tuning of gene expression or installation of novel metabolic pathways is crucial for the production of various chemicals on a large scale.8,98Also, the production of various chemicals by cyanobacterial metabolic chassis can be designed in such a way that produced chemicals are secreted into the surrounding medium.99 The development of cyanobacterial strains for the production of chemicals requires the integration of information obtained from basic physiological studies into modern-day studies such as biochemistry, molecular biology, synthetic biology, metabolic engineering, and systems biology. As depicted in Fig. 5, the developments in genetic tools, computational modeling, and high-throughput screening and sequencing techniques are continuously enhancing our capacity to engineer cyanobacteria for a variety of sustainable biotechnological purposes.100–102
Recent advancements in synthetic biology tools have facilitated the development of intricate metabolic engineering programs. A wide range of synthetic biology and metabolic engineering tools have already been developed for cyanobacteria.98 These methods include utilization of the commonly used isopropyl-β-D-thiogalactopyranoside (IPTG)-controllable trc promoter, native and synthetic promoters, riboswitches, ribosome binding sites, selectable markers, suitable vectors for stable chromosome integration, dynamic regulation of gene expression, and genome-wide editing.88,103,104
The availability of cyanobacterial genome sequencing data has enabled the integration of transcriptomics, proteomics, and metabolomics studies, leading to the development of genome-scale models for various species such as Synechocystis sp. PCC 6803, Synechococcus sp. PCC 7002, Synechococcus elongatus PCC 7942, Anabaena sp. PCC 7120, and Arthrospira platensis NIES-39.105–107 Additionally, synthetic biology tools such as CRISPR/Cas9-mediated gene editing have been successfully applied in cyanobacteria for precise modifications of the genome.108,109S. elongatus PCC 7942 was one of the first cyanobacteria, along with Synechocystis sp. PCC 6803, Anabaena sp. PCC 7120, and the fast-growing Synechococcus elongatus UTEX 2973 strain, where CRISPR editing has been successfully used.108,109 However, CRISPR/Cas9-mediated gene editing in cyanobacteria, which is otherwise well established for other organisms, is still lagging behind due to the availability of more reliable and efficient homologous recombination-based approaches for genome editing using conjugation, natural transformation, or electroporation.
8.1 Green chemistry using genetically engineered strains of cyanobacteria
It is fascinating that S. elongatus PCC 7942 and Synechocystis sp. PCC 6803 have been successfully engineered for the production of energy-rich chemicals.69,110 As mentioned above, the production of ethanol by cyanobacteria was for the first time achieved by installing two genes of a Gram-negative anaerobic bacterium Zymomonas mobilis.111 Also, cellulose production in cyanobacterium Synechococcus sp. leopoliensis strain UTCC 100 through incorporation of the cellulose synthase encoding gene from Gluconobacter xylinus provides an alternative feedstock for ethanol production.112Another four-carbon alcohol, i.e., butanol (C4H10O), has been produced through microbial fermentation and from petrochemical propylene feedstock.113–115 Butanol and isobutanol are not naturally produced by cyanobacteria; however, genetic engineering has enabled the modification of cyanobacteria Synechocystis sp. PCC 6803 and S. elongatus PCC 7942 to produce these alcohols.108,116–118 The production of these energy-rich molecules by cyanobacteria has been achieved by introducing the genes from bacteria Clostridium acetobutylicum, Treponema denticola, and E. coli.108,118,119
The production of isoprene, which is another valuable organic compound, has been accomplished by introducing the gene encoding isoprene synthase from the kudzu vine into Synechocystis sp. PCC 6803.120 Likewise, the production of ethylene in Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803 has been successfully achieved using their genetically engineered strains.121,122 Once ethylene is produced by cyanobacteria, a combination of biological and chemical conversion processes can be employed to transform ethylene into a range of hydrocarbons and functional groups that can be directly used as biofuels or as an intermediate for biofuel production.123 Thus, metabolic/genetic engineering offers potential means for utilizing renewable cyanobacterial feedstock to produce sustainable alternatives to conventional petroleum-based fuels using captured CO2.
In the cyanobacterium Synechocystis sp. PCC 6803, the production of isoprene was enhanced by introducing exogenous prokaryotic mevalonic acid (MVA) pathway genes.124 Limonene, a cyclic monoterpene present in citrus fruit rinds, has various commercial uses in the flavoring and pharmaceutical industries. It is also a potential candidate for blending into jet fuel.125 Limonene has been successfully produced using the cyanobacterium Anabaena sp. PCC 7120 by overexpressing key enzymes, i.e., the limonene synthase gene from Schizonepeta tenuifolia and three endogenous terpene biosynthesis genes of Anabaena sp. PCC 7120.123 Caffeic acid, a natural phenylpropanoid with anticancer properties, was produced in the cyanobacterium Synechocystis sp. PCC 6803 by heterologous expression of a gene from Arabidopsis thaliana.126
The genetically engineered strains of cyanobacteria have also been used to produce various valuable compounds such as glycogen, mannitol, L-lactic acid, and D-lactic acid that are promising feedstocks for the medical, pharmaceutical, chemical, food, and biofuel industries.127,128 Cyanobacteria naturally accumulate glycogen, which can be used as a feedstock for biofuel production.129,130 Under specific conditions of high-light intensity, CO2 concentration, nitrogen depletion, and moderate salinity, Synechococcus sp. PCC 7002 can produce 3.5 g per L glycogen with a productivity of 0.5 g per L per day.131 The same organism has also been engineered to produce mannitol from CO2 by overexpressing mannitol-1-phosphate dehydrogenase (MtlD) and mannitol-1-phosphatase (Mlp) encoding genes of E. coli and Eimeria tenella, respectively. The engineered strain can produce 1.1 g L−1 of mannitol with a productivity of 0.15 g per L per day.132
L-Lactic acid used in various industries can be produced from pyruvate using an NADH-dependent lactate dehydrogenase (Ldh) from Lactococcus lactis.133 The optimization strategies such as increased NADH-dependent lactate dehydrogenase (ldh) gene dosage and overexpression of the Enterococcus faecalis pyruvate kinase gene in Synechocystis sp. PCC 6803 has resulted in 840 mg per L L-lactic acid with a productivity of 22 mg per g dry cell weight per h.134–136D-Lactic acid, which is valuable for biodegradable plastics, can be produced by overexpressing the D-lactate dehydrogenase (D-Ldh) encoding gene. Overexpression of native and heterologous D-Ldh from Lactobacillus delbrueckii and inactivation of native poly-3-hydroxybutyrate and acetate pathways have resulted in an increased production of D-lactic acid in Synechocystis sp. PCC 6803.137,138
The production of 3-hydroxypropionic acid (3HP) and poly-3-hydroxybutyrate-co-4-hydroxybutyrate (P3HB4HB) has also been achieved in engineered strains of cyanobacteria S. elongatus PCC 7942 and Synechocystis sp. PCC 6803. In S. elongatus PCC 7942, two pathways, namely malonyl-CoA and β-alanine dependent pathways, for 3HP production can be engineered.139 However, in Synechocystis sp. PCC 6803, a different pathway has been engineered to directly reduce malonyl-CoA into 3HP by the overexpression of the malonyl-CoA reductase encoding gene of Chloroflexus aurantiacus.140 In S. elongatus PCC 7002, P3HB4HB production has been achieved by expressing a heterologous poly-3HB synthesis operon (phaABEC operon of Chlorogloeopsis fritschii PCC 9212). The genetically engineered strain gives the production of approximately 4.5% P3HB4HB, containing 12% 4-hydroxybutyrate (4HB), of total dry cell weight.141 Thus, genetic modification of wild-type cyanobacterial strains demonstrates the potential of these prokaryotes to be used as a host to produce biodegradable polymers using atmospheric CO2.
Cyanobacteria produce a variety of secondary metabolites, including peptides, alkaloids, and polyketides, exhibiting a wide range of biological activities.8 These metabolites are potential pharmaceuticals that can address the need for new drugs amid increasing antibiotic resistance and emerging diseases. Cyanobacterial peptides such as microcystins, nodularins, cylindrospermopsin, and cyanopeptolins show potent inhibitory activity against specific protein phosphatases and serine proteases.8 Therefore, these chemicals pose significant health risks to humans and animals. However, their specificity offers the potential for developing targeted therapies with fewer side effects in comparison to traditional drugs. These metabolites also contribute significantly to scientific research, particularly in understanding cell signaling and cancer biology.142,143 Thus, cyanobacterial toxins are valuable candidates for drug development and hold considerable potential for biotechnological applications in medicine and agriculture.
Other cyanobacterial compounds exhibit diverse biological activities such as antibacterial, antifungal, antimalarial, antiprotozoal, and antiviral.144 Also, a few cyanobacteria show antitumor activity and antimitotic activity by inhibiting microtubule assembly and colchicine binding to tubulin.145 Thus, cyanobacterial metabolites can be used to develop new pharmaceuticals such as anticancerous agents to address some of the most challenging health issues of the present day. The ongoing research and discovery of new compounds from cyanobacteria could further lead to the development of novel therapeutics with targeted mechanisms of action. Also, the continued exploration and characterization of cyanobacterial metabolites will be crucial in harnessing their full potential for medical advancement in the future.
Cyanobacterial toxins also have commercial applications as algaecides, herbicides, and insecticides. For example, the indole alkaloid hapalindole A exhibits antialgal and antimycotic properties, while norharmane from Nodularia harveyana shows anticyanobacterial activity.146 Cyanobacterin, produced by Scytonema sp., is a potent PSII inhibitor, and it is effective against algal and cyanobacterial blooms at low concentrations.147 Similarly, cyanobacterins LU-1 and LU-2 from Nostoc linckia inhibit PSII-dependent electron transport. Other notable secondary metabolites that are effective against algal blooms are nostocyclamide, nostocine A, and nostocarboline from Nostoc species.148 Additionally, pentacyclic calothrixins from Calothrix sp. inhibit transcription and DNA synthesis, while microcystin from Microcystis aeruginosa acts as a growth inhibitor for several aquatic plants.148 Also, cyanobacteria are rich sources of photoprotective compounds, mycosporine-like amino acids (MAAs) and scytonemins.8 These compounds are crucial for their application in the pharmaceutical, cosmetic, and toiletries industries due to their stability under different photophysical and photochemical conditions.149
In summary, while the primary focus on cyanobacterial toxins has been on their detrimental effects, there is growing interest in their potential advantages. These include their application in medical research and their ecological significance in aquatic systems. Therefore, further understanding the dual aspects of cyanobacterial toxins could lead to innovative approaches in both environmental management and biomedical science. This will also help in transforming a natural hazard into a resource for medical and environmental advancement.
9 Conclusions and future directions
The potential of cyanobacterial metabolic chassis to sequester CO2 and convert it into valuable chemicals is immense, and genetically engineered strains can be designed for direct secretion of synthesized chemicals into the medium. For instance, as a proof-of-concept, Ducat et al. (2012)150 developed an engineered strain of S. elongatus PCC 7942 that can produce and secrete sucrose into the growth medium. The produced sucrose can be directly harvested from the liquid medium without the need for cell harvesting and downstream processing, or it can also be used to establish a consortium between cyanobacteria and heterotrophic organisms such as yeast or eubacteria. Thus, a consortium between cyanobacteria and heterotrophic organisms opens doors for sustainable biotechnological solutions where heterotrophic organisms can thrive without needing an external carbon source. However, the success of such a consortium relies on harnessing cyanobacterial metabolic networks to either replace or enhance current production systems for specific products. Fortunately, advancements in bioprospecting and the development of platform technologies have significantly expanded our understanding of cyanobacterial metabolism to enable precise manipulation and optimization for desired outcomes. These studies underscore the potential for cyanobacteria to play a pivotal role in sustainable bioproduction and, therefore, offer commercially viable alternatives to traditional methods.One key advantage of cyanobacteria lies in their highly efficient CCM that enables them to effectively utilize bicarbonate and overcome the limitations posed by the slow diffusion of CO2 into water. By enhancing the efficiency of the cyanobacterial CCM, future work should aim to increase the concentration of CO2 around the enzyme Rubisco to improve photosynthetic efficiency and reduction of photorespiration in cyanobacterial strains targeted for chemical production. For example, an extra bicarbonate transporter has been installed in Synechocystis PCC 6803 which leads to a 2-fold enhancement in the growth rate and a higher biomass accumulation.151 Continued research into optimizing the cyanobacterial CCM and exploring novel applications in carbon-capture technologies will be crucial in combating climate change and achieving a more sustainable future. By further exploring their diverse applications, advancement in genetic engineering techniques, and scaling up their production processes, cyanobacteria can play a pivotal role in shaping a more sustainable and environment-conscious future. Therefore, continued interdisciplinary research efforts and collaborations are essential in unlocking the full potential of cyanobacteria to address global challenges and foster a greener tomorrow.
The advancements in understanding and manipulating cyanobacterial metabolism coupled with the development of new platform technologies have significantly expanded our ability to harness these organisms for sustainable bioproduction. By combining the right tools with the right cyanobacterial strain, we can achieve viable productivity and pave the way for the commercialization of cyanobacterium-based biotechnological solutions. Looking ahead, the future of cyanobacterium research and application presents exciting opportunities for innovation and impact. By harnessing the genetic and metabolic potential of these microorganisms, researchers can further enhance biofuel and food production, CO2 sequestration, and specific chemical production to achieve sustainable developmental goals in a greener way. In conclusion, cyanobacteria have immense potential to tackle global concerns related to the sustainability of food, energy, and the environment; however, the economic viability of such attempts poses a major challenge. Therefore, governments need to subsidize the costs associated with cyanobacterial and algal cultivation to achieve a carbon-neutral environment.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
PP, DP, and SPS conceptualized the idea. PP, DP, AG, RG, ST, and SPS wrote the manuscript. RG and ST prepared figures. SPS edited the manuscript. All authors reviewed the manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the Science and Engineering Research Board (SERB), New Delhi (SCP/2022/000201). SPS acknowledges support from the Institute of Eminence Incentive Grant, Banaras Hindu University (R/Dev/D/IOE/Incentive/2021-2022/32399). PP is thankful to the CSIR, New Delhi (File No.: 09/0013(15476)/2022-EMR-I). DP is thankful to the Science and Engineering Research Board (SERB), New Delhi (SCP/2022/000201), for providing a junior research fellowship. AG thanks ICMR, New Delhi, for a senior research fellowship (3/1/3/JRF-2019/HRD-LS). RG (NTA Ref. No.-211610075296) and ST (NTA Ref. No.-201610202339) are thankful to UGC, New Delhi, India, for a junior research fellowship.References
- United Nations Population Fund (UNFPA), State of World Population Report, 2024, https://www.unfpa.org.
- United Nations Population Fund (UNFPA), State of World Population Report, 2023, https://www.unfpa.org.
- Intergovernmental Panel on Climate Change (IPCC), Climate Change 2022: Impacts, Adaptation and Vulnerability, 2022, https://www.ipcc.ch.
- B. C. O'neill, M. Oppenheimer, R. Warren, S. Hallegatte, R. E. Kopp, H. O. Pörtner, R. Scholes, J. Birkmann, W. Foden, R. Licker and G. Yohe, Nat. Clim. Change, 2017, 7, 28–37 CrossRef.
- M. Barnett, Manage. Sci., 2023, 69, 7562–7584 CrossRef.
- Intergovernmental Panel on Climate Change (IPCC), Climate Change 2023: Synthesis Report, 2023, https://www.ipcc.ch.
- P. K. Maurya, S. Mondal, V. Kumar and S. P. Singh, Environ. Sci. Pollut. Res. Int., 2021, 28, 49327–49342 CrossRef PubMed.
- R. Rajneesh, S. P. Singh, J. Pathak and R. P. Sinha, Renewable Sustainable Energy Rev., 2017, 69, 578–595 CrossRef.
- J. Pathak, H. Ahmed, S. P. Singh, D.-P. Häder and R. P. Sinha, in Aquatic Ecosystems in a Changing Climate, CRC Press, 2018, pp. 45–61 Search PubMed.
- J. Zhou, H. Meng, W. Zhang and Y. Li, Adv. Exp. Med. Biol., 2018, 1080, 97–116 CrossRef PubMed.
- P. Pandey, R. Gupta, S. Tiwari, A. Gupta, S. Mondal, R. P. Sinha and S. P. Singh, in Cyanobacteria: Metabolism to Molecules, Academic Press, 2024, pp 57–67 Search PubMed.
- G. D. Price, M. R. Badger, F. J. Woodger and B. M. Long, J. Exp. Bot., 2008, 59, 1441–1461 CrossRef PubMed.
- M. R. Badger and G. D. Price, J. Exp. Bot., 2003, 54, 609–622 CrossRef.
- P. Agarwal, R. Soni, P. Kaur, A. Madan, R. Mishra, J. Pandey, S. Singh and G. Singh, Front. Microbiol., 2022, 13, 939347 CrossRef PubMed.
- S. Lucius and M. Hagemann, Front. Plant Sci., 2024, 15, 1417680 CrossRef.
- S. Mazard, A. Penesyan, M. Ostrowski, I. T. Paulsen and S. Egan, Mar. Drugs, 2016, 14, 97 CrossRef.
- D. C. Ducat, J. C. Way and P. A. Silver, Trends Biotechnol., 2011, 29, 95–103 CrossRef PubMed.
- M. S. Massey and J. G. Davis, Nitrogen, 2023, 4, 253–262 CrossRef.
- J. Pathak, R. Rajneesh, P. K. Maurya, S. P. Singh, D.-P. Häder and R. P. Sinha, Front. Environ. Sci., 2018, 6, 7 CrossRef.
- L. Curatti, M. D. Nascimento, L. A. Pagnussat, L. Sanchez Rizza, A. O. Sanchez, L. Garcia Martinez and J. A. Hernandez, Rev. Environ. Sci. Biotechnol., 2024, 23, 291–320 CrossRef.
- K. Senthilraja, S. Venkatesan, D. Udhaya Nandhini, M. Dhasarathan, B. Prabha, K. Boomiraj, S. Mohan Kumar, K. Bhuvaneswari, M. Raveendran and V. Geethalakshmi, Agriculture, 2023, 13, 1037 CrossRef.
- R. J. Cicerone and R. S. Oremland, Global Biogeochem. Cycles, 1988, 2, 299–327 CrossRef.
- J. Hu, M. Bettembourg, L. Xue, R. Hu, A. Schnürer, C. Sun, Y. Jin and J. F. Sundström, Sci. Total Environ., 2024, 920, 170980 CrossRef PubMed.
- P. L. Bodelier, Nature, 2015, 523, 534–535 CrossRef.
- S. P. Cuellar-Bermudez, J. A. Magdalena, K. Muylaert and C. Gonzalez-Fernandez, Algal Res., 2019, 44, 101689 CrossRef.
- R. Ambrosio, L. S. Rizza, M. D. Nascimento, H. G. J. Pacheco, L. M. M. Ramos, J. A. Hernandez and L. Curatti, in Cyanobacterial Lifestyle and its Applications in Biotechnology, Academic Press, 2022, pp. 99–158 Search PubMed.
- S. Tiwari, C. Singh and J. S. Singh, Energy, Ecol. Environ., 2018, 3, 355–371 CrossRef.
- J. S. Singh, A. Kumar, A. N. Rai and D. P. Singh, Front. Microbiol., 2016, 7, 529 Search PubMed.
- D. R. Nayak, Y. J. Babu, A. Datta and T. K. Adhya, J. Environ. Qual., 2007, 36, 1577–1584 CrossRef.
- R. Prasanna, V. Kumar, S. Kumar, A. K. Yadav, U. Tripathi, A. K. Singh, M. C. Jain, P. Gupta, P. K. Singh and N. Sethunathan, Microbiol. Res., 2002, 157, 1–6 CrossRef.
- L. Brennan and P. Owende, Renewable Sustainable Energy Rev., 2010, 14, 557–577 CrossRef.
- Z. Chi, J. V. O'Fallon and S. Chen, Trends Biotechnol., 2011, 29, 537–541 CrossRef PubMed.
- M. G. De Morais and J. A. V. Costa, J. Biotechnol., 2007, 129, 439–445 CrossRef.
- B. Wang, Y. Li, N. Wu and C. Q. Lan, Appl. Microbiol. Biotechnol., 2008, 79, 707–718 CrossRef PubMed.
- L. Doron and C. A. Kerfeld, Biochem. Soc. Trans., 2024, 52, 997–1010 CrossRef PubMed.
- A. Burlacot, O. Dao, P. Auroy, S. Cuiné, Y. Li-Beisson and G. Peltier, Nature, 2022, 605, 366–371 CrossRef.
- B. L. Montgomery, S. Lechno-Yossef and C. A. Kerfeld, J. Exp. Bot., 2016, 67, 2931–2940 CrossRef PubMed.
- L. Whitehead, B. M. Long, G. D. Price and M. R. Badger, Plant Physiol., 2014, 165, 398–411 CrossRef.
- J. Wolfenden and T. A. Mansfield, Proc. R. Soc. Edinburgh, Sect. B:Biol. Sci., 1990, 97, 117–138 Search PubMed.
- I. Y. López-Pacheco, L. I. Rodas-Zuluaga, S. Fuentes-Tristan, C. Castillo-Zacarías, J. E. Sosa-Hernández, D. Barceló, H. M. Iqbal and R. Parra-Saldívar, J. CO2 Util., 2021, 53, 101704 CrossRef.
- M. Anjos, B. D. Fernandes, A. A. Vicente, J. A. Teixeira and G. Dragone, Bioresour. Technol., 2013, 139, 149–154 CrossRef PubMed.
- V. Senatore, A. Buonerba, T. Zarra, G. Oliva, V. Belgiorno, J. Boguniewicz-Zablocka and V. Naddeo, Chemosphere, 2021, 273, 129682 CrossRef.
- D. J. Farrelly, C. D. Everard, C. C. Fagan and K. P. McDonnell, Renewable Sustainable Energy Rev., 2013, 21, 712–727 CrossRef.
- B. Zhao and Y. Su, Algal Res., 2020, 51, 102066 CrossRef.
- A. L. Gonçalves, Appl. Sci., 2021, 11, 871 CrossRef.
- J. Bao, C. Zhuo, D. Zhang, Y. Li, F. Hu, H. Li, Z. Su, Y. Liang and H. He, Plant Soil, 2021, 463, 97–112 CrossRef.
- B. K. Chikkaswamy, Int. J. Adv. Res. Eng. Appl. Sci., 2015, 4, 1–15 Search PubMed.
- S. H. Marzouk, H. J. Tindwa, N. A. Amuri and J. M. Semoka, Heliyon, 2023, 9, e13040 CrossRef PubMed.
- D. Chittora, M. Meena, T. Barupal and P. Swapnil P, Biochem. Biophys. Rep., 2020, 22, 100737 Search PubMed.
- J. U. Itelima, W. J. Bang, I. A. Onyimba, M. D. Sila and O. J. Egbere, Direct Res. J. Agric. Food Sci., 2018, 6, 73–83 Search PubMed.
- G. Berg, C. Zachow, H. Müller, J. Philipps and R. Tilcher, Agronomy, 2013, 3, 648–656 CrossRef.
- B. Kour, P. Sharma, S. Ramya, S. Gawdiya, K. Sudheer and B. Ramakrishnan, J. Appl. Phycol., 2024, 36, 1859–1874 CrossRef CAS.
- International Energy Agency (IEA), World energy outlook 2023, Paris, 2023, https://www.iea.org.
- B. B. Uzoejinwa, X. He, S. Wang, A. E. F. Abomohra, Y. Hu and Q. Wang, Energy Convers. Manag., 2018, 163, 468–492 CrossRef CAS.
- T. G. Ambaye, M. Vaccari, A. Bonilla-Petriciolet, S. Prasad, E. D. van Hullebusch and S. Rtimi, J. Environ. Manage., 2021, 290, 112627 CrossRef CAS PubMed.
- D. Lee, H. Nam, M. W. Seo, S. H. Lee, D. Tokmurzin, S. Wang and Y. K. Park, Chem. Eng. J., 2022, 447, 137501 CrossRef CAS.
- J. H. Hwang, J. Church, S. J. Lee, J. Park and W. H. Lee, Environ. Eng. Sci., 2016, 33, 882–897 CrossRef CAS.
- R. A. Voloshin, M. V. Rodionova, S. K. Zharmukhamedov, T. N. Veziroglu and S. I. Allakhverdiev, Int. J. Hydrogen Energy, 2016, 41, 17257–17273 CrossRef CAS.
- D. Neupane, Bioengineering, 2022, 10, 29 CrossRef PubMed.
- M. F. Demirbas, Appl. Energy, 2009, 86, S151–S161 CrossRef.
- S. Joshi and S. Mishra, Bioresour. Technol., 2022, 352, 127037 CrossRef CAS PubMed.
- S. J. Malode, K. K. Prabhu, R. J. Mascarenhas, N. P. Shetti and T. M. Aminabhavi, Energy Convers. Manag., 2021, 10, 100070 CrossRef CAS.
- S. Hayashida, K. Ohta, P. Q. Flor, N. Nanri and I. Miyahara, Agric. Biol. Chem., 1982, 46, 1947–1950 CAS.
- L. Qin, X. Zhao, W. C. Li, J. Q. Zhu, L. Liu, B. Z. Li and Y. J. Yuan, Biotechnol. Biofuels, 2018, 11, 1–10 CrossRef PubMed.
- X. H. Hu, M. H. Wang, T. Tan, J. R. Li, H. Yang, L. Leach, R. M. Zhang and Z. Luo, Genetics, 2007, 175, 1479–1487 CrossRef.
- R. A. Sheldon, ACS Sustain. Chem. Eng., 2018, 6, 32–48 CrossRef.
- W. H. Chen, K. T. Lee and H. C. Ong, Energies, 2019, 12, 290 CrossRef.
- A. Klanchui, N. Raethong, P. Prommeenate, W. Vongsangnak and A. Meechai, in Network Biology, Springer, Cham, 2017, pp. 75–102 Search PubMed.
- J. Dexter and P. Fu, Energy Environ. Sci., 2009, 2, 857–864 RSC.
- M. Wang, G. Luan and X. Lu, J. Biotechnol., 2020, 317, 1–4 CrossRef PubMed.
- P. Farrokh, M. Sheikhpour, A. Kasaeian, H. Asadi and R. Bavandi, Biotechnol. Prog., 2019, 35, e2835 CrossRef PubMed.
- A. M. P. Anahas and G. Muralitharan, Energy Convers. Manag., 2018, 157, 423–437 Search PubMed.
- F. Andrews, M. Faulkner, H. S. Toogood and N. S. Scrutton, Biotechnol. Biofuels, 2021, 14, 240 CrossRef PubMed.
- L. Cepoi, N. Donţu, V. Şalaru and V. Şalaru, in Cyanobacteria for Bioremediation of Wastewaters, Springer, Cham, 2016, pp. 27–43 Search PubMed.
- M. Danouche, N. El Ghachtouli and H. El Arroussi, Heliyon, 2021, 7, e07609 CrossRef PubMed.
- Y. Jia, W. Chen, Y. Zuo, L. Lin and L. Song, Sci. Total Environ., 2018, 613, 1324–1330 CrossRef PubMed.
- T. Sun, H. Huo, Y. Zhang, Y. Xie, Y. Li, K. Pan, F. Zhang, J. Liu, Y. Tong, W. Zhang and L. Chen, ACS Nano, 2024, 18, 17694–17706 Search PubMed.
- H. I. El Shimi and S. S. Mostafa, ARPN J. Eng. Appl. Sci., 2016, 11, 10259–10272 Search PubMed.
- A. Christodoulou and K. Stamatelatou, Water Sci. Technol., 2016, 73, 453–462 CrossRef PubMed.
- C. M. Monteiro, P. M. Castro and F. X. Malcata, Biotechnol. Prog., 2012, 28, 299–311 CrossRef CAS PubMed.
- D. Galinytė, G. Balčiūnaitė-Murzienė, J. Karosienė, D. Morudov, R. Naginienė, D. Baranauskienė, J. Šulinskienė, I. Kudlinskienė, A. Savickas and N. Savickienė, Plants, 2023, 12, 3150 CrossRef.
- S. Shanab, A. Essa and E. Shalaby, Plant Signal. Behav., 2012, 7, 392–399 CrossRef CAS.
- K. B. Chekroun, E. Sánchez and M. Baghour, Int. J. Environ. Res. Publ. Health, 2014, 1, 19–32 Search PubMed.
- Z. Zahra, Z. Habib, S. Hyun and M. Sajid, Sustainability, 2022, 14, 2041 CrossRef CAS.
- H. E. S. Touliabah, M. M. El-Sheekh, M. M. Ismail and H. El-Kassas, Molecules, 2022, 27, 1141 CrossRef CAS.
- I. Z. Ahmad, Lett. Appl. Microbiol., 2022, 75, 718–730 CrossRef CAS.
- A. C. Albertsson, C. Barenstedt and S. Karlsson, Acta Polym., 1994, 45, 97–103 CrossRef CAS.
- P. Sarmah and J. Rout, Environ. Sci. Pollut. Res. Int., 2018, 25, 33508–33520 CrossRef CAS.
- P. K. Roy, M. Hakkarainen, I. K. Varma and A. C. Albertsson, Environ. Sci. Technol., 2011, 45, 4217–4227 CrossRef CAS PubMed.
- R. V. Kumar, G. R. Kanna and S. Elumalai, J. Biorem. Biodegrad., 2017, 8, 2 Search PubMed.
- J. A. V. Costa, J. B. Moreira, B. F. Lucas, V. D. S. Braga, A. P. A. Cassuriaga and M. G. D. Morais, Ind. Biotechnol., 2018, 14, 249–256 CrossRef.
- M. Das and S. K. Maiti, J. Environ. Chem. Eng., 2021, 9, 105379 CrossRef.
- S. Ansari and T. Fatma, PLoS One, 2016, 11, e0158168 CrossRef.
- K. Meixner, I. Fritz, C. Daffert, K. Markl, W. Fuchs and B. Drosg, J. Biotechnol., 2016, 240, 61–67 CrossRef.
- E. Rueda, E. Gonzalez-Flo, S. Mondal, K. Forchhammer, D. M. Arias, K. Ludwig, B. Drosg, I. Fritz, C. R. Gonzalez-Esquer, S. Pacheco and J. García, Rev. Environ. Sci. Biotechnol., 2024, 23, 321–350 CrossRef.
- T. Lopez-Arenas, M. González-Contreras, O. Anaya-Reza and M. Sales-Cruz, Comput. Chem. Eng., 2017, 107, 140–150 CrossRef.
- D. Tiwari, N. Kumar, R. Bongirwar and P. Shukla, World J. Microbiol. Biotechnol., 2024, 40, 263 CrossRef PubMed.
- B. M. Berla, R. Saha, C. M. Immethun, C. D. Maranas, T. S. Moon and H. B. Pakrasi, Front. Microbiol., 2013, 4, 246 Search PubMed.
- S. G. Hays and D. C. Ducat, Photosynth. Res., 2015, 123, 285–295 CrossRef.
- M. Baunach, A. Guljamow, M. Miguel-Gordo and E. Dittmann, Nat. Prod. Rep., 2024, 41, 347–369 RSC.
- Y. Jeong, S. H. Cho, H. Lee, H. K. Choi, D. M. Kim, C. G. Lee, S. Cho and B. K. Cho, Microorganisms, 2020, 8, 1849 CrossRef.
- J. Nogales, S. Gudmundsson and I. Thiele, Bioengineered, 2013, 4, 158–163 CrossRef PubMed.
- G. Ma, H. Pei, W. Hu, X. Xu, C. Ma and X. Li, Bioresour. Technol., 2014, 165, 191–198 CrossRef PubMed.
- D. Camsund and P. Lindblad, Front. Bioeng. Biotechnol., 2014, 2, 40 Search PubMed.
- T. Fujisawa, R. Narikawa, S. I. Maeda, S. Watanabe, Y. Kanesaki, K. Kobayashi, J. Nomata, M. Hanaoka, M. Watanabe, S. Ehira and E. Suzuki, Nucleic Acids Res., 2017, 45, D551–D554 Search PubMed.
- Y. Kato, K. Inabe, R. Hidese, A. Kondo and T. Hasunuma, Bioresour. Technol., 2022, 344, 126196 Search PubMed.
- P. Pachauri, P. Rai and S. Srivastava, in Methods in Cyanobacterial Research, CRC Press, 2024, pp. 187–204 Search PubMed.
- X. Li, C. R. Shen and J. C. Liao, Photosynth. Res., 2014, 120, 301–310 CrossRef PubMed.
- J. Ungerer and H. B. Pakrasi, Sci. Rep., 2016, 6, 39681 CrossRef CAS PubMed.
- Y. N. Choi and J. M. Park, Bioresour. Technol., 2016, 213, 54–57 CrossRef CAS PubMed.
- M. D. Deng and J. R. Coleman, Appl. Environ. Microbiol., 1999, 65, 523–528 CrossRef CAS PubMed.
- D. R. Nobles and R. M. Brown, Cellulose, 2008, 15, 691–701 Search PubMed.
- L. Liu, Y. Wang, N. Wang, X. Chen, B. Li, J. Shi and X. Li, Biochem. Eng. J., 2021, 173, 108070 Search PubMed.
- E. L. Lan and J. C. Liao, Metab. Eng., 2011, 13, 353–363 Search PubMed.
- R. Miao, X. Liu, E. Englund, P. Lindberg and P. Lindblad, Metab. Eng. Commun., 2017, 5, 45–53 CrossRef PubMed.
- X. Liu, H. Xie, S. Roussou and P. Lindblad, Curr. Opin. Biotechnol., 2022, 73, 143–150 CrossRef CAS.
- H. Xie, J. Kjellström and P. Lindblad, Biotechnol. Biofuels Bioprod., 2023, 16, 134 Search PubMed.
- H. Luo, Y. Shi, F. Xie, T. Zhou, L. Gao, R. Yang and Z. Wang, Ind. Crops Prod., 2023, 191, 115976 CrossRef CAS.
- A. M. Varman, Y. Xiao, H. B. Pakrasi and Y. J. Tang, Appl. Environ. Microbiol., 2013, 79, 908–914 CrossRef CAS PubMed.
- P. Lindberg, S. Park and A. Melis, Metab. Eng., 2010, 12, 70–79 CrossRef CAS PubMed.
- J. Ungerer, L. Tao, M. Davis, M. Ghirardi, P. C. Maness and J. Yu, Energy Environ. Sci., 2012, 5, 8998–9006 RSC.
- C. Durall, P. Lindberg, J. Yu and P. Lindblad, Biotechnol. Biofuels, 2020, 13, 1–13 Search PubMed.
- J. N. Markham, L. Tao, R. Davis, N. Voulis, L. T. Angenent, J. Ungerer and J. Yu, Green Chem., 2016, 18, 6266–6281 Search PubMed.
- F. K. Bentley, A. Zurbriggen and A. Melis, Mol. Plant, 2014, 7, 71–86 CrossRef PubMed.
- H. Kiyota, Y. Okuda, M. Ito, M. Y. Hirai and M. Ikeuchi, Biotechnol. J., 2014, 185, 1–7 CrossRef.
- Y. Xue, Y. Zhang, S. Grace and Q. He, J. Appl. Phycol., 2014, 26, 219–226 CrossRef.
- E. Żymańczyk-Duda, S. O. Samson, M. Brzezińska-Rodak and M. Klimek-Ochab, Microorganisms, 2022, 10, 2318 CrossRef PubMed.
- Y. Kato, K. Inabe, Y. Haraguchi, T. Shimizu, A. Kondo and T. Hasunuma, Sci. Rep., 2023, 13, 7249 CrossRef PubMed.
- S. Díaz-Troya, L. López-Maury, A. M. Sánchez-Riego, M. Roldán and F. J. Florencio, Mol. Plant, 2014, 7, 87–100 CrossRef PubMed.
- A. Badary, S. Takamatsu, M. Nakajima, S. Ferri, P. Lindblad and K. Sode, Mar. Biotechnol., 2018, 20, 109–117 CrossRef.
- S. Aikawa, A. Nishida, S. H. Ho, J. S. Chang, T. Hasunuma and A. Kondo, Biotechnol. Biofuels, 2014, 7, 1–8 CrossRef PubMed.
- J. H. Jacobsen and N. U. Frigaard, Metab. Eng., 2014, 21, 60–70 CrossRef PubMed.
- A. Joseph, S. Aikawa, K. Sasaki, Y. Tsuge, F. Matsuda, T. Tanaka and A. Kondo, Biosci. Biotechnol. Biochem., 2013, 77, 966–970 CrossRef PubMed.
- S. A. Angermayr and K. J. Hellingwerf, J. Phys. Chem. B, 2013, 117, 11169–11175 CrossRef.
- S. A. Angermayr, A. G. Rovira and K. J. Hellingwerf, Trends Biotechnol., 2015, 33, 352–361 Search PubMed.
- A. D. Van der Woude, S. A. Angermayr, V. P. Veetil, A. Osnato and K. J. Hellingwerf, J. Biotechnol., 2014, 184, 100–102 CrossRef CAS.
- S. Zhou, Y. Shao, N. Gao, L. Li, J. Deng, M. Zhu and S. Zhu, Sci. Total Environ., 2014, 482, 208–213 CrossRef PubMed.
- I. S. Huang and P. V. Zimba, Harmful Algae, 2019, 86, 139–209 CrossRef PubMed.
- Y. Wang, T. Sun, X. Gao, M. Shi, L. Wu, L. Chen and W. Zhang, Metab. Eng., 2016, 34, 60–70 CrossRef PubMed.
- M. Hügler, C. Menendez, H. Schägger and G. Fuchs, J. Bacteriol., 2002, 184, 2404–2410 CrossRef.
- S. Zhang, Y. Liu and D. A. Bryant, Metab. Eng., 2015, 32, 174–183 CrossRef PubMed.
- R. Carpine and S. Sieber, Curr. Res. Biotechnol., 2021, 3, 65–81 CrossRef.
- T. Nawaz, L. Gu, S. Fahad, S. Saud, Z. Jiang, S. Hassan, M. T. Harrison, K. Liu, M. A. Khan, H. Liu and K. El-Kahtany, Food Energy Secur., 2023, 12, e495 CrossRef.
- R. A. D. Al-Nedawe and Z. N. B. Yusof, Microb. Bioact., 2023, 6, 1–16 CrossRef.
- A. Bouyahya, S. Bakrim, I. Chamkhi, D. Taha, N. El Omari, N. El Mneyiy, N. El Hachlafi, M. El-Shazly, A. Khalid, A. N. Abdalla and K. W. Goh, Biomed. Pharmacother., 2024, 170, 115989 CrossRef.
- A. Holland and S. Kinnear, Mar. Drugs, 2013, 11, 2239–2258 CrossRef PubMed.
- J. P. Berry, M. Gantar, M. H. Perez, G. Berry and F. Noriega, Mar. Drugs, 2008, 6, 117–146 CrossRef.
- G. Zanchett and E. C. Oliveira-Filho, Toxins, 2013, 5, 1896–1917 CrossRef PubMed.
- R. P. Rastogi and A. Incharoensakdi, Photochem. Photobiol. Sci., 2014, 13, 1016–1024 CrossRef PubMed.
- D. C. Ducat, J. A. Avelar-Rivas, J. C. Way and P. A. Silver, Appl. Environ. Microbiol., 2012, 78, 2660–2668 CrossRef PubMed.
- N. A. Kamennaya, S. Ahn, H. Park, R. Bartal, K. A. Sasaki, H. Y. Holman and C. Jansson, in Metabolic Engineering, Elsevier, 2015, pp.76–85 Search PubMed.
Footnote |
| † Joint first authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2025 |