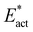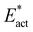The activating capture of N2 at the active site of Mo–nitrogenase†
Ian
Dance

School of Chemistry, UNSW Sydney, NSW 2052, Australia. E-mail: i.dance@unsw.edu.au
First published on 8th August 2024
Abstract
Dinitrogen is inherently inert. This report describes detailed density functional calculations (with a 485+ atom model) of mechanistic steps by which the enzyme nitrogenase activates unreactive N2 at the intact active site FeMo-co, to form a key intermediate with bound HNNH. This mechanism does not bind N2 first and then add H atoms, but rather captures N2 (‘N2-ready’) that diffuses in through the substrate channel and enters a strategic gallery of H atom donors in the reaction zone, between Fe2 and Fe6. This occurs at the E4 stage of the complete mechanism. Exploration of possible reactions of N2 in this space leads to the conclusion that the first reaction step is transfer of H on Fe7 to one end of N2-ready, soon followed by Fe–N bond formation, and then a second H transfer from bridging S2BH to the other N. Two H–N bonds and one or two N–Fe bonds are formed, in some cases with a single transition state. The variable positions and orientations of N2-ready lead to various reaction trajectories and products. The favourable products resulting from this capture, judged by the criteria of reaction energies, reaction barriers, and mechanistic competence for further hydrogenation reactions in the nitrogenase cycle, have Fe2–NH–NH bonding. The trajectory of one N2 capture reaction is described in detail, and calculations that separate the H atom component and the ‘heavy atom’ components of the classical activation energy are described, in the context of possible H atom tunneling in the activation of N2-ready. I present arguments for the activation of N2 by the pathway of concerted hydrogenation and binding of N2-ready, alternative to the commonly assumed pathway of binding N2 first, with subsequent hydrogenation. The active site of nitrogenase is well primed for the thermodynamic and kinetic advantages of N2 capture.
1. Introduction
The conversion of N2 to NH3 is essential for all life on earth. The microbial enzyme nitrogenase is the natural catalyst for this process, but now roughly half of the food for human life is fertilised by ammonia from the industrial Haber–Bosch process,1 which is very energy expensive and emits unacceptable amounts of CO2.2 Concerns about the consequences for the earth's climate from all greenhouse gas emissions, and the transition away from fossil fuels, have generated new attention on NH3, produced from N2, as a practical carrier for H2 as a fuel, or in NH3 fuel cells.3–6The hydrogenation of N2 to NH3 is thermodynamically favourable,  NH3 (g) = −3.9 kcal mol−1. However the reaction pathway from N2 to NH3 is kinetically difficult. The bond dissociation energy of N2 is 225 kcal mol−1. The remarkable unreactivity of the N2 molecule is also due to its non-polarity, and the large gap of 10.8 eV between the highest occupied and lowest unoccupied molecular orbitals.7 The hydrogenation of N2 to diazene N2H2 is endothermic, +49.6 kcal mol−1.8 Other fundamental properties related to the initial hydrogenated species from N2 are collected in ref. 8. Emphatic expression of the fact that the kinetic difficulty in converting N2 to NH3 occurs at the first hydrogenation step appears in the initially large negative standard reduction potentials plotted in Fig. 1.
NH3 (g) = −3.9 kcal mol−1. However the reaction pathway from N2 to NH3 is kinetically difficult. The bond dissociation energy of N2 is 225 kcal mol−1. The remarkable unreactivity of the N2 molecule is also due to its non-polarity, and the large gap of 10.8 eV between the highest occupied and lowest unoccupied molecular orbitals.7 The hydrogenation of N2 to diazene N2H2 is endothermic, +49.6 kcal mol−1.8 Other fundamental properties related to the initial hydrogenated species from N2 are collected in ref. 8. Emphatic expression of the fact that the kinetic difficulty in converting N2 to NH3 occurs at the first hydrogenation step appears in the initially large negative standard reduction potentials plotted in Fig. 1.
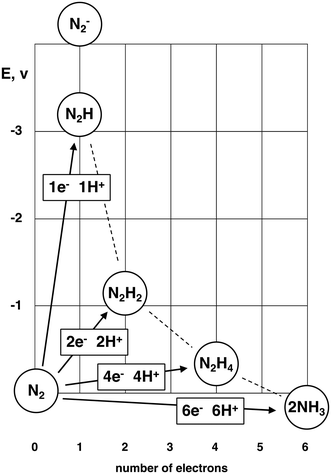 | ||
| Fig. 1 Standard reduction potentials (vs. NHE) for reduction of N2. From refs. 8–11. | ||
Clearly any mechanism for the hydrogenation of N2 needs to account for the first step to N2H. The enzyme nitrogenase hydrogenates N2 to NH3 with relative ease under ambient conditions, and so must have an effective pathway for the first hydrogenation of N2. I report here investigations of the mechanism of these intrinsically difficult first steps for the activation of N2 as they are effected by the enzyme nitrogenase.
The catalytically active site that has evolved to effect this chemistry is a multi-metal cluster, CFe7MoS9, the largest metallocluster of enzymology.12,13Fig. 2a shows this cluster, which together with an obligatory homocitrate ligand (HCA), is known as the iron-molybdenum cofactor, FeMo-co. It is located in the MoFe protein that together with the Fe protein comprises the nitrogenase enzyme.14,15 At the centre of the cluster is a six-coordinate C atom (Cc), bonded to a trigonal prism of Fe atoms (Fe2 to Fe7).16–19 Three S atoms doubly-bridge the axial edges of this trigonal prism, and at each end of the prism a set of three triply-bridging S atoms connect to Fe1 or Mo. There are only two bonds to the surrounding protein, through cysteine coordination of Fe1, and histidine coordination at Mo. Each Fe atom is four-coordinate, approximately tetrahedral, while Mo is octahedrally coordinated. The mechanistically influential amino acids that surround FeMo-co, identified by experimental investigations of the consequences of amino acid mutations on the enzyme reactions and reactivity,15,20–34 are shown in Fig. 2b: throughout this paper amino acids are numbered according to crystal structure PDB 3U7Q of species Azotobacter vinelandii (Av). Val70 and the side chain of Arg96 flank the active face and NεH of His195 forms a good hydrogen bond to S2B.
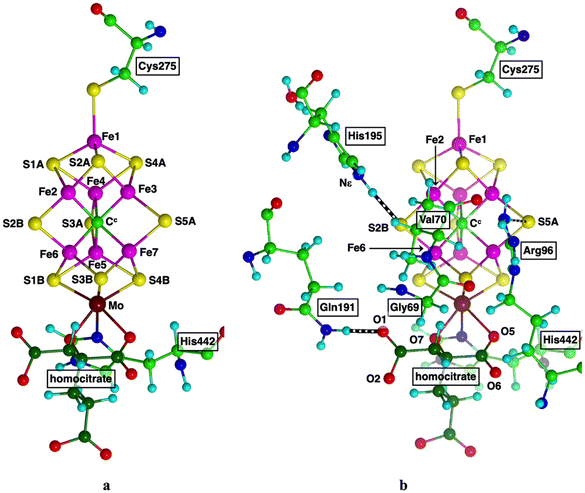 | ||
| Fig. 2 (a) Structure of FeMo-co, with connections via Cys275, His442. Labels for Av1 protein, crystal PDB 3U7Q. Homocitrate C atoms are dark green. (b) Significant surrounding amino acids. Hydrogen bonds are striped. | ||
Mutagen-reactivity experiments show that the active domain of FeMo-co is the front face, Fe2–S2B–Fe6–S3B–Fe7 enclosed by Val70 and Arg96 (see Fig. 2b).20–22,26,30,31,33–37 Assumptions that this domain remains intact during the catalytic cycle have been challenged in recent years, by crystal structures and reactivities of nitrogenase isoforms in which S2B has been displaced into the protein by another atom or ligand.38–46 The possible lability of S2B has attracted computational investigations.47–55 An alternative hemilability of S2B, in which only one of the S2B–Fe bonds is broken during catalysis, has been explored computationally.50,56–62 In the results presented here the S2B–Fe2 and S2B–Fe6 bonds remain, and FeMo-co is intact.
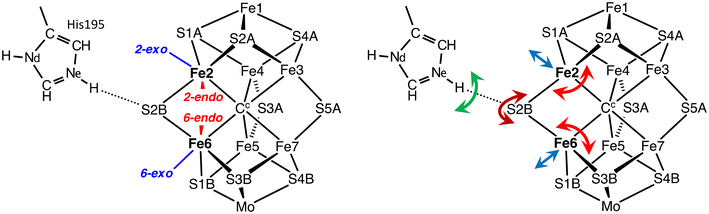
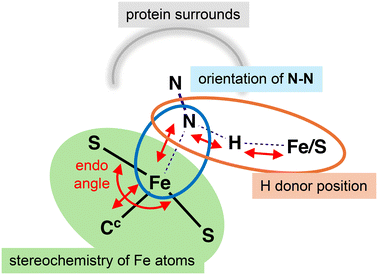
In the FeMo-co resting state the central Fe atoms are four coordinate and pseudo-tetrahedral. Ligands can bond to the exo and endo coordination positions, shown on Fig. 3a for Fe2 and Fe6, and this bonding induces modification of the FeMo-co cluster core as marked by the arrows on Fig. 3b. The coordination stereochemistry at ligated Fe can be tetrahedral, trigonal bipyramidal, or octahedral.63,64Exo ligation of Fe usually causes extension of the trans Fe–Cc bond (blue arrows). For endo coordination (and endo plus exo coordination) the geometrical adjustment is more substantial, because the S2B–Fe2–S2A and S2B–Fe6–S3B angles (the ‘endo angles’) expand from near trigonal (120°) in the unligated cluster to nearer 180° (red arrows). Changes in the endo angles are associated with a folding backwards or forwards of S2B (violet arrows) Fig. 3b, a movement that is connected to hydrogen bonding between Nε of His195 and S2B. The side chain of His195 can move substantially in several directions (green arrows) in conjunction with this plasticity of FeMo-co. These distortions of the FeMo-co core incur adaptation energy penalties.65
1.1 Background
Two fundamental components of the initial hydrogenation of N2 are the location and mode of N2 binding, and the sources of H atoms. Note that these H atoms may be polarized slightly positive or slightly negative, according to their bonded atoms: they are neither hydride ions H− nor protons H+, both of which cannot exist as such on FeMo-co. In this paper H represents a slightly polarised H atom on Fe or S, or an acidic H on Nε of His195 or on carboxylate or coordinated OH of HCA. ‘Proton transfer’ describes movement of a positively polarised H atom. The H atoms are generated by the introduction of protons along the water chain, as previously described in detail,66 concomitant with electron addition to FeMo-co.67I have previously described the ingress of N2, along a well-defined channel in the protein leading directly to the exocoordination position of Fe2.68 This pathway has been characterised by the structures of crystals pressurised with Xe69 and molecular dynamics calculations.70 N2 can bind end-on (η1) at exo-Fe2, with a binding energy that becomes more favourable as the degree of hydrogenation of FeMo-co increases.68 Calculated binding energies in the range −7 to −14 kcal mol−1 and association barriers ranging 2 to 7 kcal mol−1 have been reported, and the entropy penalty for the binding of N2 from the N2 pathway is estimated to be 4 kcal mol−1.65 However, the prospects for hydrogenation of this exo-Fe2–N–N group are not favourable. Possible H donors are HisH+, S2B–H, or endo-Fe2–H, but as shown previously these are very awkward stereochemically.68 This unreactive N2 at the exo-Fe2 position is proposed71 to be the N2 required to enable the non-physiological HD reaction, D2 + 2H + + 2e− → 2HD.72
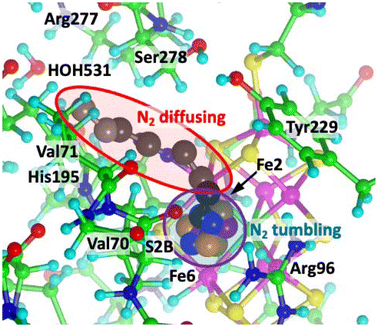
A second N2 molecule can enter along the N2 ingress pathway, past exo-Fe2–N2, and, without interference, enter the reaction zone between Fe2 and Fe6.68,73 The red enclosure of Fig. 4 shows a sequence of diffusing positions for this second N2. These positions are equi-energetic within 3 kcal mol−1, and are sustained by van der Waals interactions with non-polar H atoms of adjacent residues (mainly Ser278, Val70 and Val71) at distances ranging upwards from 2.35 Å and in the stabilising range of the N⋯H dispersion energy potential. There are no contacts less than 3 Å between the first exo-Fe2–N2 and the second incoming N2. In this reaction zone this second N2 can tumble freely: the blue enclosure of Fig. 4 shows (in different colours) four tumbling positions in the reaction cavity enclosed by Fe2, S2B, Fe6, Val70, Arg96, Fe3 and Fe7. The energy profile as N2 diffuses in and tumbles is relatively flat.
1.2 Premises for N2 capture at the E4 stage
The mechanistic cycle of nitrogenase is commonly described with the scheme developed by Thorneley and Lowe (TL) from seminal early kinetic measurements.74 This model has been analysed further75 and elucidated with a recent analysis of new steady-state kinetic data.76 The TL scheme involves a sequence of En states, generated by addition of electron + proton pairs. The TL scheme describes the important relationships between N2 and H2 because exogenous H2 is a competitive inhibitor of the reduction of N272,74,77 and this is embodied in the N2/H2 exchange equilibria of the TL mechanism.Exchange of H2 and N2 and binding of N2 occur at the E4 stage of the TL mechanism. In a complete mechanism (E0 to E8) to be described separately a necessary component is the accumulation of five added protons with four added electrons at E4. Although it is generally assumed that proton addition is coupled with electron addition, there is no experimental evidence for equal numbers of added protons and electrons at each stage. The E4 5H stage (i.e. 4 added electrons, 5 added protons) of the full mechanism includes H2 generated and bound at the endo position of Fe6, and it is this H2 that dissociates to expose endo-Fe6 for N2 capture in the reaction zone. The reason for the additional proton at the E4 stage arises later in the mechanism, which uses this proton at a stage where its replenishment by migration is still blocked.78
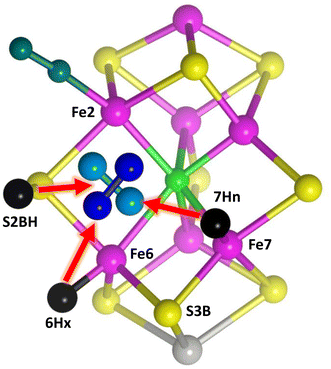
The starting point for the involvement of N2 is the arrangement shown in Fig. 5. Following previous surveys of the accumulation of H atoms on FeMo-co in the earlier stages of the mechanism,64,79 there are H atoms on S2B (S2BH), at exo-Fe6 (6Hx) and at endo-Fe7 (7Hn). S2B is the most stable location for H on FeMo-co,64,79,80exo-Fe6–H also has high stability, and 7Hn has been reported as on the path of incoming protons.55 H2 has just dissociated from endo-Fe6 in the H2/N2 exchange,78 allowing the reducible N2 to have various positions and orientations in the reaction cavity. Two postulated orientations of N2 are shown in Fig. 5: in the following these are described as N2-ready-a (light blue) and N2-ready-b (dark blue). A crucial feature of the premised scenario in Fig. 5 is that three H atoms are primed (red arrows) to capture N2 with H–N bond formation, concurrent with N2 forming bond(s) with Fe2 and/or Fe6. This general array in Fig. 5 is stereochemically propitious for the crucial first hydrogenation and binding of N2, and is the starting point for the investigations described here.
2. Methodology
2.1 Protein model
The computed protein model is a 485+ atom extract from crystal 3U7Q, including all relevant amino acids. This is my standard model for simulations of nitrogenase reactions and reactivity.65,71,73,81 Details, and the rationale for inclusion of amino acids and truncation of uninvolved side chains, are provided in the ESI.† Investigations of the protonation state of homocitrate using crystal structures at various pH (summarised in ref. 82), together with vibrational circular dichroism spectroscopy,83 QM/MM calculations,82,84 and quantum refinement,84 indicate that the coordinated alcoholate O7 atom is protonated and hydrogen bonded to O1, and this is now included in the computed model. Some constraints on the protein structure are required during optimization calculations because the modelled protein is incomplete and the influences of the complete protein outside the computational model are absent. The modelled protein must also maintain flexibility sufficient to accommodate the coordination of N2 and of H2, and the diffusion of these molecules in the vicinity of FeMo-co. The constraining strategy and the 28 distance constraints are described in the ESI.† In the reaction zone the distances Cα(Val70)–Cc and Cα(Val70)–CZ(Arg96) are fixed. The charge on the [CFe7MoS9] core of FeMo-co is −1, in agreement with experimental and computational studies.81,82,85,862.2 Density functional procedures
Density functional (DF) calculations use the DMol methodology of Delley,87–92 with accurate DNP (double numerical plus polarisation) basis sets.90 The gradient-corrected functional PBE93 is used because validation tests demonstrate that when used with the numerical basis sets of DMol it is more accurate than other commonly used functionals.94 See the ESI† for validation information. The conductor-like screening model (COSMO)95–97 is used with a dielectric constant of 5. The dispersion components of the non-bonding intermolecular interactions are treated effectively with the numerical basis sets used here: see the ESI† for additional information. Constraints on interatomic distances use the Lagrange Multiplier Algorithm. Control of electronic states is via input specifications of spin populations for Fe1, Fe3, Fe4, Fe5 and Fe7, subsequently optimised.2.3 Electronic states for the structures reported here
The electronic states of the structures reported here are characterised by the spin densities on the Fe atoms, and the total spin S. I have previously shown that the electronic states with negative spin densities on Fe3 and Fe5 (labelled ‘35’), or on Fe4 and Fe7 (‘47’), are generally the most stable,68 and this is consistent with the findings and procedures of other authors.54,60,82,98–104 The rationale for this stability of the 35 and 47 electronic states is published.68 These two electronic states were used in the present calculations, together with total spins S = 1/2 and S = 3/2. In general the structures and reactions reported here were calculated in the four combinations of electronic and spin state, labeled as 35(1/2), 35(3/2), 47(1/2), 47(3/2).2.4 Mapping potential energy surfaces
This investigation comprehensively mapped the potential energy topology in the reaction space of Fig. 5, in order to locate energy minima, and then to locate the energy saddle points between them. Reactant N2 is unbound, and there are multiple geometrical variables, identified generally in Fig. 6. In addition to the position and orientation of the reactant N2, there are variable positions for the H donors (H on Fe or S), and the core of FeMo-co is plastic. The Fe2–S2B–Fe6 bridge is intact in all structures, and all N–Fe bond formation occurs at the endo positions of Fe2 and/or Fe6. Fe–N ligation changes the endo angles at Fe (S2B–Fe2–S2A, S2B–Fe6–S3B), and the coupled Cc–Fe distances. All reported models have the same number of atoms and electrons, so energies are directly comparable.The exploration deployed many calculations of energy minimisation trajectories, starting with the possible locations and orientations of N2, and positions of the H atoms on S2B and at endo-Fe7, guided always by the relevant stereochemical principles. The shape of the potential energy surface was monitored throughout, and the magnitude of the displacement in each energy minimisation step was adjusted to maintain regular small energy changes.
Transition states and intrinsic reaction coordinates were determined by the procedure described previously66,81,105,106 and explained in detail in the ESI.† Because there are multiple variables in the systems described here (see Fig. 6) including descriptors of the orientation of unbound N–N, the TS calculations are less straightforward than for conventional bond breaking and making reactions. Some reactions contain more than one bond breaking-bond making step. Application of the relevant principles of stereochemistry is important for these manual calculations of reaction trajectories and transition states. There are variations of Fe spin densities during most reactions, and continuity of the electronic state throughout each reaction trajectory is checked.
2.5 Activation energy analysis
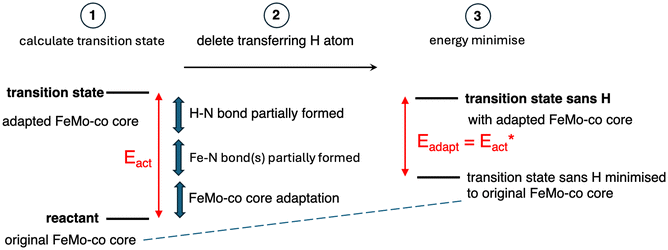
The reactions described here involve H transfer reactions, which might occur, at least in part, with H atom tunneling. Hydrogen tunneling, a quantum mechanical phenomenon, is possible when there is overlap of the probability functions of the reactant and product states. The effect of tunneling can be regarded as a lowering of the barrier of classical transition state theory. Tunneling is often evaluated in terms of the de Broglie wavelength, which for a hydrogen atom with ambient (300 K) thermal energy of 2.5 kcal mol−1 is ca. 0.5A: the concept is that as the H atom ascends a barrier it can tunnel through it when the barrier width is of similar dimension.107In view of the possibility of H atom tunneling, additional calculations were made to separate the components of the activation potential energy that involve movements of non-H atoms (‘heavy atoms’), and the components which primarily involve H atom movements. This is not a calculation of a tunneling correction, which is beyond the capability of the present calculations. The concept here is that removal of the H atom from a calculated structure removes its contribution to the calculated energy, leaving the heavy atom components. The computational procedure is illustrated in Scheme 1. The calculated activation energy, Eact has contributions from the formation of the H–N bond, the formation of N–Fe bond(s), and the adaptation of the cluster core (Fig. 6). Here, the H atom component of the energy barrier is separated by the following calculation protocol. The transferring H atom is deleted from the transition state structure, and the energy of the ‘transition state sans H’ is minimised until the FeMo-co core and any Fe–N interactions revert to their dimensions in the reactant structure. These changes, and their limit, are evident primarily in the Fe–Cc distances and endo angles at Fe2, Fe6, Fe7. This energy change, Eadapt, is the activation energy devoid of the H–N bond formation components and is labelled 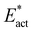 in the following. The adaptation energy calculated from ‘transition state sans H’ necessarily has total spin S different from that of the transition state, and that difference was maintained as ±1/2. The necessarily different electronic structure of ‘TS-sans-H’ relative to the TS causes an energy adjustment at the outset of the core reversion energy minimisation, prior to the geometry changes during the minimisation, and, by using miniscule displacements initially, this was carefully separated from the energy changes as the core geometry reverts. The component of the reaction barrier that involves H atom movements is
in the following. The adaptation energy calculated from ‘transition state sans H’ necessarily has total spin S different from that of the transition state, and that difference was maintained as ±1/2. The necessarily different electronic structure of ‘TS-sans-H’ relative to the TS causes an energy adjustment at the outset of the core reversion energy minimisation, prior to the geometry changes during the minimisation, and, by using miniscule displacements initially, this was carefully separated from the energy changes as the core geometry reverts. The component of the reaction barrier that involves H atom movements is 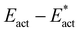 . There is some uncertainty in identifying the minimisation limit in the calculation of Eadapt and values have a precision of 1–2 kcal mol−1.
. There is some uncertainty in identifying the minimisation limit in the calculation of Eadapt and values have a precision of 1–2 kcal mol−1.
3. Results
The reaction space of Fig. 5 was comprehensively explored, starting with possible locations and orientations of N2, and of the H donors, to find stable structures in which N2, N2H or N2H2 is bonded to Fe2 and/or Fe6. I will first describe the scope of structures found and investigated, together with some pictures showing dimensions, followed by tabulation of relative energies. I will then describe the reactions that form and transform these structures, with their potential energy profiles and activation barriers Eact and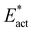 . Analysis of the reaction trajectories and components of the potential energy barriers leads to considerations of H atom tunnelling in the initial stages. Then, the products of these reactions, with bound HNNH, are assessed against the criterion of mechanistic competence.
. Analysis of the reaction trajectories and components of the potential energy barriers leads to considerations of H atom tunnelling in the initial stages. Then, the products of these reactions, with bound HNNH, are assessed against the criterion of mechanistic competence.
3.1 Scope
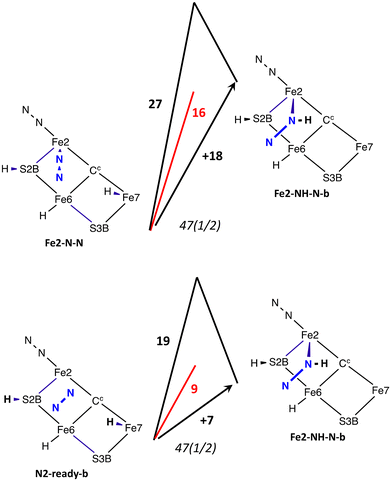
The structures found and investigated are portrayed as minimal skeletons in Scheme 2, which also defines their labels. There are two principal orientations of unbound N2, N2-ready-a and N2-ready-b. These are not arbitrary positions but are obtained as the reactant limit in reaction trajectories from transition states, as described below. A third orientation, N2-ready-c, is part of only one trajectory.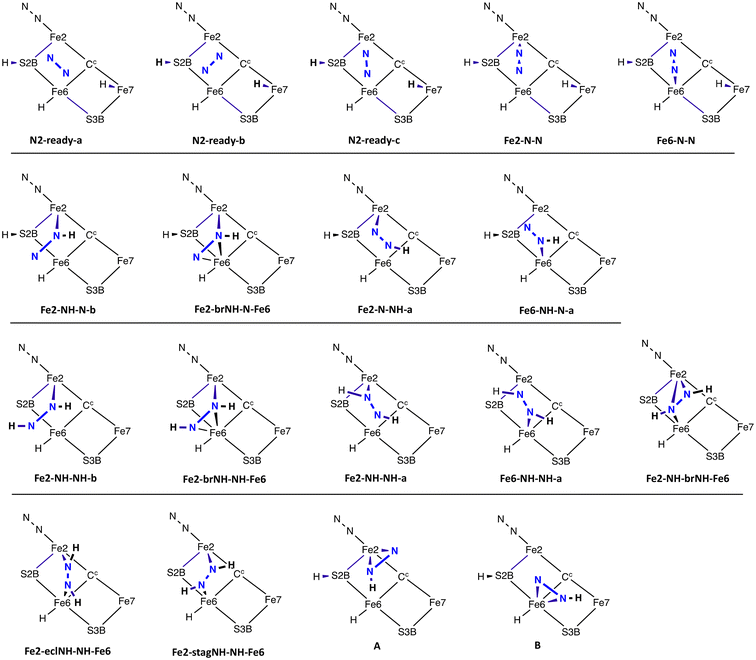 | ||
| Scheme 2 The structures investigated and their labels. Suffixes a and b define the oblique orientation of the N–N bond (as in Fig. 5). ‘br’ in the label signifies bridging Fe2 and Fe6, ‘ecl’ signifies the eclipsed conformation of cis diazene, and ‘stag’ the staggered conformation. | ||
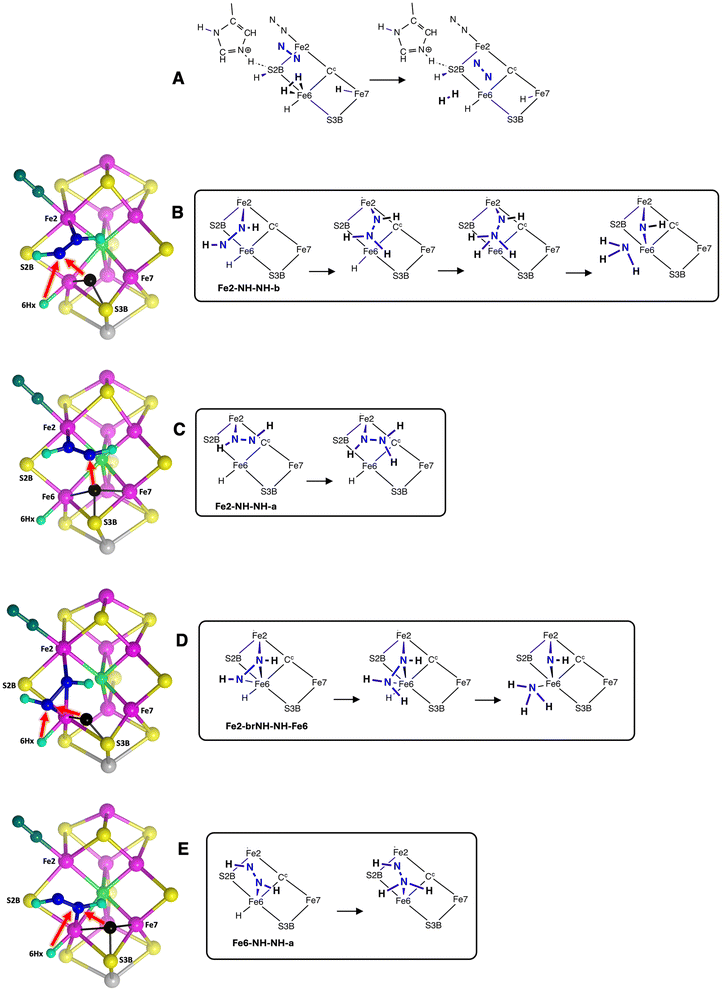
(a) Two obvious structures have N2 bound end-on (η1) at the endo coordination positions of Fe2 (Fe2-N-N) or Fe6 (Fe6-N-N).
(b) Structures with one N–H bond are the second set of four in Scheme 2. Three stable structures contain NNH bound to Fe: they are Fe2-NH-N-b and Fe2-N-NH-a with one Fe–N bond, and Fe2-brNH-N-Fe6 in which the NNH ligand bridges Fe2 and Fe6. In these structures the H atom is sourced from Fe7–H, and S2BH is unchanged. The Fe6 analog of Fe2-NH-N-b, Fe6-NH-N-a, in not an energy minimum and undergoes barrierless transfer of H from S2B, forming Fe6-NH-NH-a.
(c) Five structures contain bound diazine HNNH. They are Fe2-NH-NH-b, Fe2-NH-NH-a and Fe6-NH-NH-a with HNNH bound to one Fe atom, while Fe2-brNH-NH-Fe6 and Fe2-NH-brNH-Fe6 contain HNNH bridging both Fe atoms. Diazine bound end-on between Fe2 and Fe6 is stable only as the eclipsed (cis) conformation, in structure Fe2-eclNH-NH-Fe6. Diazine in the staggered conformation and bound only end-on between Fe2 and Fe6, as in Fe2-stagNH-NH-Fe6, or the alternative conformation, is unstable, transforming without barrier to Fe2-NH-brNH-Fe6 or Fe2-eclNH-NH-Fe6. The predominance of bound diazene in the trans conformation is a consequence of the positions of the Fe7H and S2BH donors. Two conceivable structures with side-on bonding of N2H (A and B in Scheme 2) are not stable.
All structures use the endo coordination positions of Fe2 or Fe6, and all contain a normal Fe2–S2B–Fe6 bridge.
Pictures which reveal core details of the minimised structures are provided in Fig. 7, with selected dimensions marked, and with the first picture showing the contextual surrounding amino acids. The N–N distances increase as expected with the degree of bonding to N–N. The N–N distances are 1.12 Å for end-on NN, 1.19–1.21 Å for bound NNH, 1.25–1.27 Å where HNNH has one bond to Fe, and 1.31–1.33 Å when HNNH has three bonds to Fe in Fe2-brNH-NH-Fe6 and Fe2-NH-brNH-Fe6 (not shown).
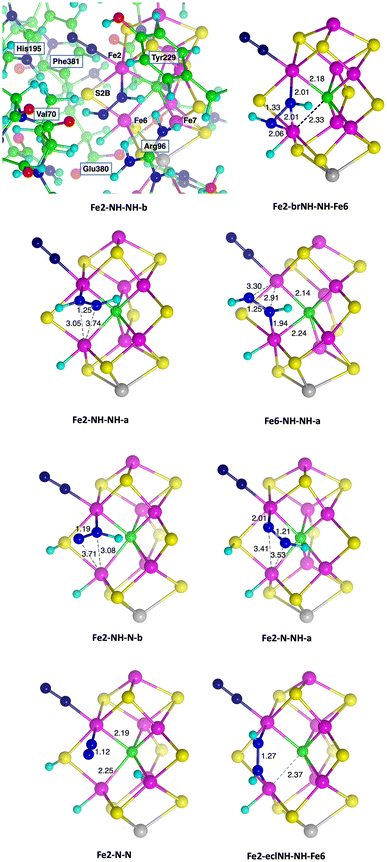 | ||
| Fig. 7 Core representations of some optimised structures, with selected dimensions (Å) marked. The top left panel includes the nearby amino acids as they occur for all structures. | ||
3.2 Energies
All of the structures described here were calculated with the standard protein structure (section 2.1) and have the same number of atoms and electrons, so that straightforward comparisons of energy are possible. Similarly, the same electronic and spin states were used for all structures. The relative energies of the found structures are presented in Table 1, and are referenced to the energy of the N2-ready-a structure with the same electronic/spin state.| Structure | S = 3/2, 35 | S = 3/2, 47 | S = 1/2, 35 | S = 1/2, 47 |
|---|---|---|---|---|
| N2-ready-a | 0 | 0 | 0 | 0 |
| N2-ready-b | 3 | 6 | 7 | 3 |
| N2-ready-c | 5 | |||
| Fe2-N-N | −9 | −10 | −10 | −6 |
| Fe6-N-N | 0 | −3 | −3 | 3 |
| Fe2-NH-N-b | 8 | 11 | 13 | 13 |
| Fe2-brNH-N-Fe6 | 14 | 14 | 17 | 17 |
| Fe2-N-NH-a | 4 | 2 | 4 | 6 |
| Fe2-NH-NH-b | −14 | −14 | −10 | −13 |
| Fe2-brNH-NH-Fe6 | −10 | −8 | −3 | −4 |
| Fe2-NH-NH-a | −15 | −18 | −13 | −15 |
| Fe6-NH-NH-a | −6 | −7 | −4 | −2 |
| Fe2-NH-brNH-Fe6 | −4 | −4 | 4 | 0 |
| Fe2-eclNH-NH-Fe6 | −15 | −16 | −12 |
Energies, here and in subsequent analyses, are given at 1 kcal mol−1 precision. This is because uncertainties of this magnitude arise in several ways. (a) N2 in the N2-ready structures is unbound and variable in position and orientation, with energy variations of ca. 1 kcal mol−1. (b) Structures are generally calculated as the limit of the trajectory from a transition state, either towards reactant or towards product. Variations of ca. 1 kcal mol−1 arise between structures calculated as reactant in one reaction trajectory and product in another.
Several generalisations can be made from the results in Table 1.
(a) The two structures with end-on Fe–N–N bonding are more stable by up to 10 kcal mol−1 than the N2-ready forms.
(b) Structures with bound N2H are unstable relative to N2-ready.
(c) Structures containing bound HNNH are more stable than those with bound N2H, by 20 to 30 kcal mol−1.
(d) Structures containing HNNH bound to Fe6 are less stable by ca. 10 kcal mol−1 than analogous structures bound to Fe2.
(e) Structures containing an NH bridge between Fe2 and Fe6 are less stable than analogous unbridged structures by 3 to 9 kcal mol−1.
(f) The most stable structures are Fe2-NH-NH-b, Fe2-NH-NH-a and Fe2-eclNH-NH-Fe6.
(g) In general the S = 3/2 state is slightly more stable than S = 1/2, but there is no systematic differentiation of the 35 and 47 electronic states.
The structures in the collection presented here need to be judged by criteria other than energy. In multi-step reaction mechanisms, as occur in nitrogenase, possible intermediates are assessed by the pathway(s) by which they are formed, and the pathway(s) by which they can react further. Therefore I present the potential energy profiles for the formation of structures with bound N2, bound N2H and bound N2H2, and the reaction profiles for additional N2 → N2H, N2 → N2H2 and N2H → N2H2 steps.
3.3 Reaction profiles
Possible reactions were profiled by a protocol that first located transition states (TS) between the various structures, then followed the trajectories from each TS to the reactant and product structures. Energy profiles for each of the reactions are presented in Fig. 8. For each reaction the result for one electronic/spin state is drawn: the full collection of results is in Table 2. As described in section 2.5 two calculations were made. One is the standard activation energy Eact and the other is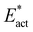 , the activation energy devoid of the contributions from the H atom transfer from Fe7 to N.
, the activation energy devoid of the contributions from the H atom transfer from Fe7 to N. 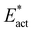 is the ‘heavy atom’ component of the activation barrier, and the difference
is the ‘heavy atom’ component of the activation barrier, and the difference 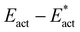 contains mainly the energy components due to H atom movements. In Fig. 8Eact is drawn black and
contains mainly the energy components due to H atom movements. In Fig. 8Eact is drawn black and 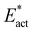 is red. Note that some reactions take N2 to HNNH with only one reaction barrier, and some have an energy minimum at the NNH intermediate. In all cases the first H transfer is from Fe7H, with the second from S2BH.
is red. Note that some reactions take N2 to HNNH with only one reaction barrier, and some have an energy minimum at the NNH intermediate. In all cases the first H transfer is from Fe7H, with the second from S2BH.
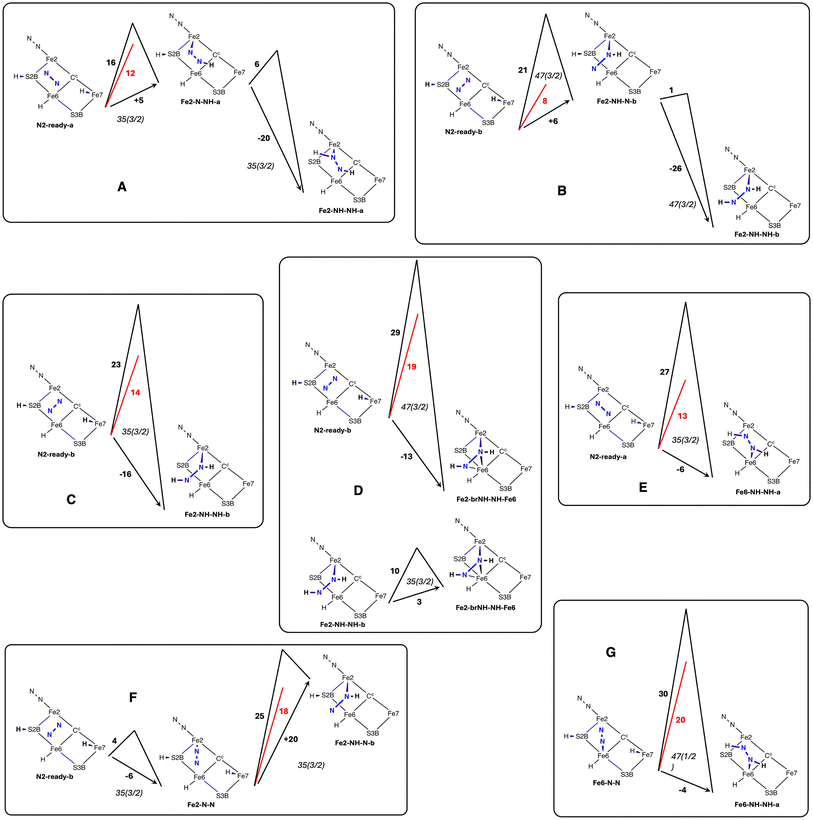 | ||
Fig. 8 Reaction energy profiles, in kcal mol−1, showing Eact in black, 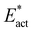 in red, and the reaction energy against the arrow. in red, and the reaction energy against the arrow. | ||
| Reaction | Electronic (spin) sate | Reaction energy | E act | |
|---|---|---|---|---|
| N2-ready-b → Fe2-NH-NH-b | 35(3/2) | −16 | 23 | 14 |
| N2-ready-b → Fe2-NH-N-b | 47(3/2) | 6 | 21 | 8 |
| 47(1/2) | 7 | 19 | 9 | |
| 35(1/2) | 10 | 22 | 6 | |
| N2-ready-b → Fe2-brNH-N-Fe6 | 35(1/2) | 11 | 29 | 21 |
| Fe2-brNH-N-Fe6 → Fe2-brNH-NH-Fe6 | 35(1/2) | −21 | <1 | |
| Fe2-NH-N-b → Fe2-NH-NH-b | 47(3/2) | −25 | <1 | |
| 47(1/2) | −27 | <1 | ||
| 35(3/2) | −22 | <1 | ||
| N2-ready-b → Fe2-brNH-NH-Fe6 | 35(3/2) | −14 | 28 | 24 |
| 47(3/2) | −14 | 29 | 19 | |
| 47(1/2) | −15 | 26 | 22 | |
| Fe2-NH-NH-b → Fe2-brNH-NH-Fe6 | 35(3/2) | 3 | 10 | |
| 47(3/2) | 5 | 11 | ||
| N2-ready-a → Fe6-NH-NH-a | 35(3/2) | −6 | 27 | 13 |
| 47(3/2) | −7 | 31 | 15 | |
| 35(1/2) | −5 | 32 | 11 | |
| 47(1/2) | −7 | 29 | 13 | |
| N2-ready-a → Fe2-N-NH-a | 35(3/2) | 5 | 16 | 12 |
| 47(3/2) | 3 | 18 | 10 | |
| 47(1/2) | 8 | 20 | 9 | |
| Fe2-N-NH-a → Fe2-NH-NH-a | 35(3/2) | −20 | 6 | 6 |
| 47(3/2) | −19 | 6 | ||
| 35(1/2) | −20 | 3 | ||
| 47(1/2) | −20 | 7 | 7 | |
| N2-ready-c → Fe2-N-N | 35(3/2) | −6 | 4 | |
| Fe2-N-N → Fe2-NH-N-b | 35(3/2) | 20 | 25 | 18 |
| 47(1/2) | 18 | 27 | 16 | |
| Fe2-N-N → Fe2-N-NH-a | 35(3/2) | 17 | 26 | 14 |
| 47(1/2) | 12 | 20 | 18 | |
| N2-ready-b → Fe6-N-N | 47(3/2) | −7 | 6 | |
| Fe6-N-N → Fe6-NH-NH-a | 35(3/2) | −6 | 32 | 21 |
| 35(1/2) | −1 | 30 | 17 | |
| 47(1/2) | −4 | 30 | 20 |
Fig. 8A shows the trajectory for N2-ready-a → Fe2-N-NH-a → Fe2-NH-NH-a in electronic state 35(3/2). The first step, transfer from Fe7–H, is 5 kcal mol−1 endergonic and then the second step, transfer from S2BH, is 20 kcal mol−1 exergonic, with a net energy change of −15 kcal mol−1. For the first step the classical activation energy Eact is 16 kcal mol−1, and 4 kcal mol−1 of this (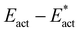 ) is attributed to H atom transfer. In the second exergonic step Eact for H atom transfer from S2B is small, 6 kcal mol−1. Fig. 8B shows a similar pair of steps, N2-ready-b → Fe2-NH-N-b → Fe2-NH-NH-b in electronic state 47(3/2). The first step is 6 kcal mol−1 endergonic, but the second is 26 kcal mol−1 exergonic, net reaction energy −19 kcal mol−1. For the first step there is some variation in the numbers for the three electronic states, 47(3/2), 47(1/2), 35(1/2) (see Table 2), but the overall dynamics are similar, Eact is 19–22 kcal mol−1,
) is attributed to H atom transfer. In the second exergonic step Eact for H atom transfer from S2B is small, 6 kcal mol−1. Fig. 8B shows a similar pair of steps, N2-ready-b → Fe2-NH-N-b → Fe2-NH-NH-b in electronic state 47(3/2). The first step is 6 kcal mol−1 endergonic, but the second is 26 kcal mol−1 exergonic, net reaction energy −19 kcal mol−1. For the first step there is some variation in the numbers for the three electronic states, 47(3/2), 47(1/2), 35(1/2) (see Table 2), but the overall dynamics are similar, Eact is 19–22 kcal mol−1, 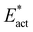 is 6 to 9 kcal mol−1. S2BH transfer to N in the second step is again easy, with an activation barrier ca. 1 kcal mol−1.
is 6 to 9 kcal mol−1. S2BH transfer to N in the second step is again easy, with an activation barrier ca. 1 kcal mol−1.
In contrast to these two instances of two-step formation of bound HNNH, the reactions in Fig. 8C, D and E capture N2-ready and form bound HNNH in a single step, and the favourable second S2BH → N transfer seen in schemes A and B is now barrierless. N2-ready-a → Fe6-NH-NH-a is exergonic by 6 kcal mol−1, N2-ready-b → Fe2-NH-NH-b is exergonic by 16 kcal mol−1, and N2-ready-b → Fe2-brNH-NH-Fe6 is exergonic by 13 kcal mol−1, values similar to the overall exergonicity of the analogous sequences in schemes A and B. The N2-ready-b → Fe2-brNH-NH-Fe6 reaction is slightly different in the 35(1/2) electronic state, because there is a very shallow (ca. 1 kcal mol−1) minimum at the Fe2-brNH-N-Fe6 intermediate (see Table 2).
Further comparisons are insightful. N2-ready-b → Fe2-NH-N-b → Fe2-NH-NH-b (Fig. 8B) has the same reactant and product as N2-ready-b → Fe2-NH-NH-b (Fig. 8C), but the reaction profiles differ because the electronic states are different. The single step N2-ready-b → Fe2-NH-NH-b in Fig. 8C occurs with electronic state 35(3/2), while the two step version in Fig. 8B occurs with electronic states 47(3/2), 47(1/2), 35(1/2). Compare also N2-ready-b → Fe2-NH-NH-b (Fig. 8C) and N2-ready-b → Fe2-brNH-NH-Fe6 (Fig. 8D). Both have the same orientation of N2-ready and the same orientation of product HNNH, but only the second involves bridging to Fe6. The different trajectories and products arise from differing positions of N2-ready at the outset. When N2-ready is central in the reaction zone the bridging product Fe2-brNH-NH-Fe6 is formed, while when incoming N2-ready is nearer to Fe2 than Fe6 the product is unbridged Fe2-NH-NH-b. The two reactions are similarly exergonic (−13, −16 kcal mol−1), but the activation barriers Eact and 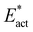 are ca. 5 kcal mol−1 larger to form the bridged product. The profile for the conversion of unbridged Fe2-NH-NH-b to bridged Fe2-brNH-NH-Fe6 is shown in Fig. 8D.
are ca. 5 kcal mol−1 larger to form the bridged product. The profile for the conversion of unbridged Fe2-NH-NH-b to bridged Fe2-brNH-NH-Fe6 is shown in Fig. 8D.
Schemes F and G in Fig. 8 show profiles for reactions involving end-on Fe–N–N. The formation of Fe2-N-N from N2-ready-b is exergonic (−6 kcal mol−1) with a small barrier of 4 kcal mol−1. The formation of Fe6-N-N is similar. The transfer of Fe7–H to Fe2-N-N yields Fe2-NH-N-b but the step is endergonic by 20 kcal mol−1 with relatively large values for Eact (27 kcal mol−1) and 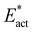 (16 kcal mol−1). For Fe6-N-N, transfer of H from Fe7 is followed in a single trajectory by H transfer from S2B, producing Fe6-NH-NH-a. The overall reaction is exergonic by 4 kcal mol−1, but the activation barriers are now considerably larger, Eact (30 kcal mol−1) and
(16 kcal mol−1). For Fe6-N-N, transfer of H from Fe7 is followed in a single trajectory by H transfer from S2B, producing Fe6-NH-NH-a. The overall reaction is exergonic by 4 kcal mol−1, but the activation barriers are now considerably larger, Eact (30 kcal mol−1) and 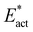 (20 kcal mol−1). These results exemplify the difficulty in hydrogenating N2 when bound end-on to Fe.
(20 kcal mol−1). These results exemplify the difficulty in hydrogenating N2 when bound end-on to Fe.
3.4 Trajectory analysis
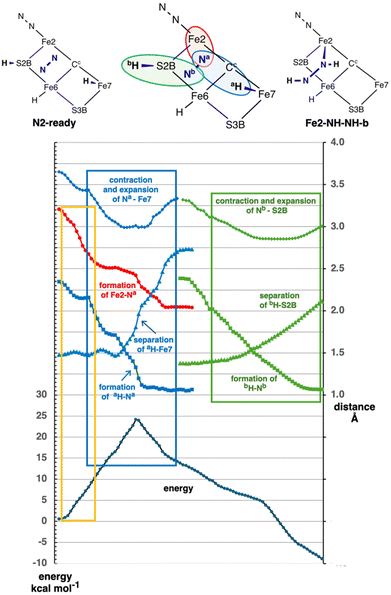
The preceding results reveal significant reactions in which N2 in an N2-ready position is captured with the formation of H–N and Fe–N bonds. These results describe reactant, transition states, and products. I now examine the course of these reactions more closely. The calculations to establish a transition state, and the pathways from transition state to reactant or product, reveal significant attributes that are now described.Fig. 9 is a detailed analysis of the trajectory for the conversion N2-ready-b to Fe2-NH-NH-b, in electronic state 35(3/2). There are sequential phases along the multivariable reaction coordinate, with one transition state. Colour differentiation of the component steps is used in Fig. 9, with abbreviations aH and Na for the first transfer, bH and Nb are the second (see cartoon). The initial section (orange enclosure of Fig. 9) involves the entrance of N2 into the reaction zone, with shortening of Fe2–Na (red) from >3 Å to a non-bonding distance of ca. 2.55 Å. At the same time Na is moving towards Fe7 (blue diamonds), although still quite distant at ca. 3.3 Å. The energy increase of ca. 12 kcal mol−1 during this approach of N2 (orange enclosure) is due in part to adaptation of the FeMo-co geometry (section 2.5).
In the next phase (blue enclosure) Fe7–Na shortens, Na–aH shortens and aH-Fe7 lengthens. This is the transfer of aH from Fe7 to Na. It occurs with a contraction of the donor–acceptor distance Fe7–Na from 3.3 to 3.0 Å during the transition. At the same time the Na is moving with low energy gradient towards Fe2, but the Fe2–Na bond does not fully form at 2.1 Å until after the aH transfer step is completed. Most of the aH transfer involves an energy climb towards the transition state, while most of the Fe2–Na bond formation occurs beyond the transition state. Note that the energy gradient is relatively steep near the transition state, because several bonding connections are changing.
The third component of the reaction (green enclosure, Fig. 9) is the second bH transfer step, from S2B to Nb. Again this H transfer step involves a contraction and subsequent expansion of the donor–acceptor distance, Nb–S2B. There is no potential energy barrier for this step, and an overall energy change of ca. −15 kcal mol−1.
The general pattern of N2-ready capture and hydrogenation pictured in Fig. 9 is characteristic of the other reactions of Fig. 8 schemes C, D and E. In all cases the energy climb to the TS involves adaptation of the FeMo-co core and the first H transfer from Fe7 to N, while the Fe–N bond formation begins near the top of the reaction barrier and completes after the TS. Variations occur mainly in the numerical values. The origin of the difference between single step transformations (schemes C, D, E) and two-step transformations (schemes A, B) is evident in the trajectory domain between the blue and green enclosures of Fig. 9, where there is smaller downhill energy gradient. In the two step reactions this part of the profile occurs as an energy minimum, which is about 1 kcal mol−1 deep in the N2-ready-b → Fe2-NH-NH [47(3/2)] reaction, and 6 kcal mol−1 deep in N2-ready-a → Fe2-NH-NH-a. A geometric factor in the differentiation of single step and two-step pathways is the partial rotational freedom around the Fe–Na bond formed in the first step.
The reaction profile shown in Fig. 9 is typical of other similar conversions. However these processes have some fluidity, and there are variations in distances and energies. There is no significant difference in trajectory between reactions that form the Fe–N bond at Fe2 or Fe6 or both (N2-ready-b → Fe2-brNH-NH-Fe6). This is because the N2-ready conformations are approximately equi-(non-bonding)-distant from these two Fe atoms. There are energy variations as shown in Fig. 8.
3.5 Mechanistic competence
In assessing a reaction step and the intermediates it connects in a catalytic mechanism cycle, an essential consideration is the feasibility of both the preceding formation step and the subsequent reaction step. There must be a way in and a way out. This is the mechanistic competence criterion. In the present context of N2 capture and hydrogenation, the pathway(s) to the capture precursor, and the subsequent reaction(s) of captured HNNH must be feasible. The reaction prior to the formation of the N2-ready precursors here is dissociation of H2 from the endo position of Fe6 as N2 diffuses in past Fe2, shown in Fig. 10A.78 This is the established H2/N2 exchange at the E4 stage of the TL nitrogenase mechanism.74,76,108–110 In my current mechanism this H2 is formed using an H atom occupying an endo position of Fe2/Fe6 or an Fe2–H–Fe6 bridge. This H atom blocks inwards diffusion of N2 into the reaction zone, and so must be removed. This is achieved by converting it to H2 (using another H from the proton supply chain). In this way dissociation of H2 is required to allow introduction and reaction of N2, explaining the H2/N2 exchange.78Next, considering subsequent steps in the mechanism, there are four structures with bound HNNH resulting from N2-capture. These are shown in Fig. 10B, C, D and E together with possible directions for transfer of the next H atom, consistent with favourable stereochemistry at N. The red arrows show how an incoming H atom (black sphere) from the proton supply chain and positioned in one of the several possible configurations near S3B, Fe6 and/or Fe755,66 could transfer to N to form a bound HNNH2 intermediate. There is no other H atom source at Fe2 or S2B that could provide this first H atom transfer to HNNH. For Fe2-NH-NH-b the first transfer from S3BH would yield Fe2–NH–NH2, and a subsequent transfer of 6Hx would yield Fe2–NH–NH3, which could then separate NH3 as shown, and is well positioned to deliver NH3 to the NH3 egress channel.111 For Fe2-brNH-NH-Fe6 (Fig. 10C) the first and second H transfers from S3BH and 6Hx are similar, but there is an alternative in which NH3 is bound to Fe6 in concert with the breaking of N–N. These steps are stereochemically competent. For Fe2-NH-NH-a the first transfer shown (Fig. 10D) yields Fe2-NH-NH2. Rotation about the Fe2–N bond might improve the stereochemistry at both N atoms for transfer of 6Hx. For the N2 capture product Fe6-NH-NH-a (Fig. 10E) a first transfer from 6Hx is possible (note that S3BH transfer is collisional), yielding Fe6-NH2-NH, but there is no feasible opportunity to further hydrogenate the distal NH, and therefore this product of N2 capture is judged to be mechanistically less competent.
The conclusion here is that the N2 capture reactions that generate Fe2-NH-NH-b, Fe2-brNH-NH-Fe6 and Fe2-NH-NH-a are mechanistically competent. In Fe2-NH-NH-a and Fe2-NH-NH-b the Fe6–N distances are about 3 Å (see Fig. 7), considerably longer than the bonds in Fe2-brNH-NH-Fe6. Further calculations are investigating these possibilities.
4. Summary
Here is a succinct summary of the main results.• Reducible N2, diffused into the reaction zone, can be positioned and oriented such that a first H atom transfer from Fe7 can cooperate with Fe–N bond formation to capture N2 as Fe bound N2H, which is then easily hydrogenated to Fe bound HNNH, either without barrier or with small barrier of ≤6 kcal mol−1.
• Endo Fe7–H is the significant donor. It is adjacent to and formed from the terminus of the proton wire supply chain.
• The S2B bridge between Fe2 and Fe6 is retained, and the H atom of S2B has a crucial role in the second hydrogenation of N2.
• The second H transfer from S2B to N is facilitated by the bending of Fe–N–N caused by the first transfer of H from Fe7.
• The array of H donors includes 6Hx, present throughout, but away from incoming N2 and unable to participate in the initial hydrogenation. It is primed for subsequent H transfer to an intermediate beyond the HNNH stage.
• Conversion of free N2 to Fe-bound HNNH is exergonic and in the range −13 to −20 kcal mol−1 for all cases except the formation of Fe6-NH-NH-a where the reaction energy is −6 kcal mol−1.
• Classical potential energy barriers for capture and hydrogenation of N2 (single step, or the first of two-step sequences) range 16 to 32 kcal mol−1. The smallest barriers, 16–20 kcal mol−1, are for the reaction N2-ready-a → Fe2-N-NH-a. The next smallest (19–23 kcal mol−1) are for N2-ready-b → Fe2-NH-N-b or Fe2-NH-NH-b. Then comes formation of the bridging intermediates, N2-ready-b → Fe2-brNH-N-Fe6 or Fe2-brNH-NH-Fe6, 26–29 kcal mol−1, and the largest occurs when the first Fe–N bonding is to Fe6, N2-ready-a → Fe6-NH-NH-a, 27 to 32 kcal mol−1.
• Intermediates in which NNH or HNNH is bridged between Fe2 and Fe6 are less stable than unbridged analogs bonded to one Fe, and have larger formation barriers.
• The energy climb to the classical transition state, Eact, contains components due to (a) H (light) atom movements, and (b) all other heavier atom movements, including adaptation of the FeMo-co cluster core geometry and possibly partial formation of Fe–N bonds. Adaptation of the cluster core is most evident as changes in the endo angles at Fe, and in the Fe–Cc distances. By deletion of H from the transition state and calculation of the energy required to revert to the reactant state, the ‘heavy’ atom component of the activation energy barrier is calculated, as Eact*. The difference, 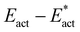 , is attributed to H atom movements that might occur with some quantum tunneling, so that actual reaction barriers could be less than Eact.
, is attributed to H atom movements that might occur with some quantum tunneling, so that actual reaction barriers could be less than Eact.
• The adaptation energy 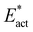 for N2 capture reactions is smallest in N2-ready-b → Fe2-NH-N-b (ranging 6 to 9 kcal mol−1 over the electronic states) and largest (19 to 24 kcal mol−1) in N2-ready-b → Fe2-brNH-NH-Fe6.
for N2 capture reactions is smallest in N2-ready-b → Fe2-NH-N-b (ranging 6 to 9 kcal mol−1 over the electronic states) and largest (19 to 24 kcal mol−1) in N2-ready-b → Fe2-brNH-NH-Fe6.
• The reactions and intermediates are calculated for the most probable electronic and spin states.
5. Discussion
The results presented here show that substrate N2 diffusing into the reaction zone of the active site can be captured by surrounding H atoms, forming HNNH bound to one or two Fe atoms. The capture of N2 to form HNNH can occur in a single step, or pass through an Fe–NNH intermediate in a shallow energy well. I discuss here the advantageous characteristics of N2 capture, the retention of the S2B bridge, and assess the reactions and products and electronic states. I consider connections of this theory to experimental information.5.1 Advantageous characteristics of N2 capture
Unbound N2 in an N2-ready position and conformation is more favourably hydrogenated than bound N2. This contravenes the conventional view of the activation of N2 by metal systems, in which the binding of N2 initiates activation, with subsequent hydrogenation.112–116 The modes of binding N2 usually considered for FeMo-co are end-on, side-on, or a bridging combination of both.65,117 End-on bonding of N2 at exo sites of Fe2 or Fe6 places the N2 away from H donors, and therefore introduces difficulties for any subsequent hydrogenation of exo bound N2.117 N2 bound end-on at the endo positions of Fe2 or Fe6 are closer to H donors, and these endo Fe2–N–N and Fe6–N–N possibilities are evaluated comparatively in this study.The differences between H transfer to unbound N2 and to bound N2 in the nitrogenase context can be seen in the comparison of two reactions (in the same electronic state) forming Fe2-NH-N-b, from Fe2-N-N and from N2-ready-b, shown in Fig. 11. The reaction potential energy from bound N2 is +18 kcal mol−1, but is +7 kcal mol−1 from unbound N2. This is because bound N2 has lower potential energy than unbound N2 (Table 1). I have previously described comprehensive calculations of the binding of the reducible N2 at the endo positions of Fe2 or Fe6, with binding potential energies relevant to the present system (i.e. intact S2B bridge) in the range −6 to −14 kcal mol−1.73 Similarly, the values in Table 1 above show that Fe–N–N structures have potential energies of −9 to −17 kcal mol−1 relative to N2-ready. Thorhallsson et al. reported comparable values of −6.9, −10.2, −13.5 kcal mol−1 for the binding of N2 end-on at exo-Fe2 or exo-Fe6.60 The entropy change asssociated with the bonding of N2 from a diffusible location within the protein is estimated to contribute about 4 kcal mol−1.65,73 It is clear that, in the context of the first hydrogenation of N2, bound N2 starts at a free energy disadvantage of ca. 10 kcal mol−1 relative to unbound N2 in an N2-ready position. This is the thermodynamic argument for the N2-ready capture method of activating N2, rather than the ‘bind first, then hydrogenate’ pathway.
Fig. 11 illustrates also the kinetic advantage of the N2-ready capture pathway, with an energy barrier of 19 kcal mol−1 compared with 27 kcal mol−1 for bound N2. Most of the difference in barrier energies is due the different relative energies of the reactants, because the energies and geometries of the two transition states are similar. The pathway from N2-ready-b starts with N–N suitably angled towards the product, but the pathway from linear Fe2–N–N requires initial bending. This difference is manifest in the larger adaptation energy, 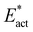 , for the Fe–N–N pathway, 16 vs. 9 kcal mol−1 (Fig. 11).
, for the Fe–N–N pathway, 16 vs. 9 kcal mol−1 (Fig. 11).
It is worth reiterating the attributes in Fig. 5: the reaction zone of nitrogenase is well able to generate and provide the gallery of nearby H donors at locations stereochemically propitious for transfer to N2-ready. These H atoms are replenishable. It is also important to recognise that there is considerable geometrical flexibility in the reactions of N2-ready. The N2 entry is fluid, FeMo-co is plastic (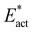 values), and the Fe–NHNH structures have some rotational freedom around the Fe–N bond. The influence of the conformational variability of N2-ready is evinced by N2-ready-b in state 47(3/2) transforming to Fe2-NH-NH-b or to Fe2-brNH-NH-b or to Fe6-N-N.
values), and the Fe–NHNH structures have some rotational freedom around the Fe–N bond. The influence of the conformational variability of N2-ready is evinced by N2-ready-b in state 47(3/2) transforming to Fe2-NH-NH-b or to Fe2-brNH-NH-b or to Fe6-N-N.
5.2 Assessment of reactions and products
Several variations of N2 capture reactions are described in this report. The criteria to assess each reaction are reaction energy, classical reaction barrier, and mechanistic competence. All reactions capturing free N2 and forming Fe–NHNH at Fe2, including the bridging form Fe2-brNH-NH-Fe6, are suitably exergonic (−13 to −20 kcal mol−1), while at Fe6 alone the formation of Fe6-NH-NH is less exergonic (−6 kcal mol−1). The classical reaction barrier criterion favours the two-step reactions, i.e. formation of Fe2-NH-NH-aviaFe2-N-NH-a and the formation of Fe2-NH-NH-bviaFe2-N-NH-b. The mechanistic competence criterion also supports Fe2-NH-NH-a and Fe2-NH-NH-b. A favourable attribute of Fe2-NH-NH-b is that the NH3 formed after two more H transfer steps (Fig. 10) is delivered to the putative mouth of the NH3 egress channel.111 The bridging intermediate Fe2-brNH-NH-Fe6, while similarly mechanistically attractive, is disfavoured slightly by the activation energies Eact and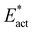 . Thus the conclusion from the present results is that Fe2-NH-NH-b is the most likely structure for the HNNH intermediate at the E4 stage of the nitrogenase mechanism. Fe2-NH-NH-b, Fe2-NH-NH-a and Fe2-brNH-NH-Fe6 are included in ongoing calculations of the next steps as suggested in Fig. 10.
. Thus the conclusion from the present results is that Fe2-NH-NH-b is the most likely structure for the HNNH intermediate at the E4 stage of the nitrogenase mechanism. Fe2-NH-NH-b, Fe2-NH-NH-a and Fe2-brNH-NH-Fe6 are included in ongoing calculations of the next steps as suggested in Fig. 10.
5.3 Electronic structures
The electronic structures of reactants and of the potential energy surfaces followed in the reactions described here have been selected as those generally accepted as possessing the best energies, from surveys54,82,98–104 and by recognition of basic principles of electronic structure for ligated FeMo-co.68 As is usually seen in the reactions of FeMo-co, the spin densities on Fe atoms undergoing ligation variation can change appreciably during the course of reaction steps. It is necessary to check that these spin density variations occur within the one electronic state being followed, and are not jumps to different electronic states. Electronic states can influence the shape of the potential energy surface and the reaction outcome: N2-ready-b in the 35(3/2) state transforms to Fe2-NH-NH-b in one step, while in the 47(3/2), 35(1/2) and 47(1/2) states passes through the shallow minimum of Fe2-NH-N-b.5.4 The S2B bridge is intact
The mechanisms presented here for the first part of the N2 hydrogenation sequence of nitrogenase include the intact S2B bridge between Fe2 and Fe6, and S2B in this bridging location is a crucial component, functioning via S2B–H as a facile H donor to the captured N2. The precise position of S2B is variable, bending backwards and forwards as the endo angles at Fe2 and Fe6 change, but the S2B–Fe2 bond and the S2B–Fe6 bond are retained throughout. There is much discussion, founded on experimental findings, about the possibility that S2B is partially or completely displaced from FeMo-co during the catalytic cycle, summarised in ref. 115 There is no experimental evidence that the physiological nitrogenase cycle involves disruption of the S2B bridge. The results presented here show that the first stages of N2 activation can occur favourably without disruption of the S2B bridge.The present results also reveal a significant and general attribute of the reactivity of the active site, focussed on the intact S2B bridge. The H transfer from S2BH to N is a facile step, aided by the proximity of S2BH to the reaction zone between Fe2 and Fe6 where the first binding of N2 occurs, a proximity that is evident in the pictures in Fig. 7. The S2B bridge is also close to the protonated Nε atom of His195, with an Nε-H⋯S2B hydrogen bond in the resting state. This allows H transfer, in both directions, between His195 and S2B. His195Nε can function as an H atom buffer, but only if S2B is in its bridging position. These properties are deployed in a complete mechanism I have developed,78 which starts with His195NεH and S2BH and includes these functions of S2B and His195Nε to provide two H atoms first to N2 (as in the present results) and then to a subsequent intermediate. After completion of the hydrogenation of N2 and the release of two NH3 the same functions of S2B and His195Nε are used to regenerate the resting state with His195NεH and S2BH.
5.5 Connections with experiment
The devising of experiments to test the N2 capture mechanistic proposals made here is challenging. How does the N2 capture hypothesis relate to existing experimental information? Seefeldt et al. devised clever experimentation to employ diazene, HNNH, as substrate.118 Diazene has a half-life of about 5s. The production of NH3 from diazene is similar to that from N2, and, significantly, is similarly inhibited by H2. This suggests that N2 and HNNH follow the same pathway early in the mechanism of nitrogenase. In the context of the present report this would be explained in terms of substrate HNNH diffusing into the reaction zone and forming one of the Fe–NHNH intermediates described here. In the absence of N2 the exo position of Fe2 would also be available to bind HNNH, but as with N2 in this position, further H transfer to exo-Fe2–HNNH would be impeded by lack of well-placed H donors. Cao and Ryde calculated numerous possibilities for HNNH bound to FeMo-co, including trans HNNH bound at this exo position of Fe2 as one of the most stable structures in their survey.119 They reported that the HNNH group is stacked between His195 and Ser278, but is not stabilised by any hydrogen bonds. This supports the view that the experimental reaction with HNNH as substrate involves binding of HNNH at exo-Fe2 but that this HNNH would not react further. A second HNNH would diffuse into the reaction cavity, and would be primed for further reaction with Fe–N bonding.6. Conclusion
This investigation provides an answer to the fundamental question raised in the introduction - how does nitrogenase avoid the inherent difficulty in the initial hydrogenation of N2? FeMo-co is able to conduct a concerted attack on N2 entering the reaction zone, using two strategically placed H atom donors, as well as Fe atoms Fe2 and Fe6. The first part of the attack is H transfer to N, before Fe–N bond formation. This initial deployment of an H transfer step introduces the possibility of some quantum tunneling of H through the initial barrier, diminishing the activation barrier and facilitating the activation of N2. In short, I propose that the inherent unreactivity of N2 is overcome by the ability of the enzyme to arrange a well-positioned gallery of H atoms for the first attack. The active site of nitrogenase is well primed for the thermodynamic and kinetic advantages of N2 capture and activation.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
I thank Professor Sam Hay and a reviewer for advice about H atom tunneling. This research was undertaken with the aid of resources from the National Computational Infrastructure (NCI Australia), a facility enabled by the Australian National Collaborative Research Infrastructure Strategy. My research is funded by UNSW Sydney.References
-
V. Smil, Enriching the Earth: Fritz Haber, Carl Bosch, and the Transformation of World Food Production, MIT Press, Cambridge, Massachusetts, 2001 Search PubMed
.
- I. Rafiqul, C. Weber, B. Lehmann and A. Voss, Energy efficiency improvements in ammonia production—perspectives and uncertainties, Energy, 2005, 30, 2487–2504 CrossRef CAS
.
- O. Siddiqui and I. Dincer, A review and comparative assessment of direct ammonia fuel cells, Therm. Sci. Eng. Prog., 2018, 5, 568–578 CrossRef
.
- G. Chehade and I. Dincer, Progress in green ammonia production as potential carbon-free fuel, Fuel, 2021, 299, 120845 CrossRef CAS
.
- P. Adamou, S. Bellomi, S. Hafeez, E. Harkou, S. M. Al-Salem, A. Villa, N. Dimitratos, G. Manos and A. Constantinou, Recent progress for hydrogen production from ammonia and hydrous hydrazine decomposition: A review on heterogeneous catalysts, Catal. Today, 2023, 423, 114022 CrossRef CAS
.
- D. S. Dhawale, S. Biswas, G. Kaur and S. Giddey, Challenges and advancement in direct ammonia solid oxide fuel cells: a review, Inorg. Chem. Front., 2023, 10, 6176–6192 RSC
.
- C.-G. Zhan, J. A. Nichols and D. A. Dixon, Ionization Potential, Electron Affinity, Electronegativity, Hardness, and Electron Excitation Energy: Molecular Properties from Density Functional Theory Orbital Energies, J. Phys. Chem. A, 2003, 107, 4184–4195 CrossRef CAS
.
- J. A. Pople and L. A. Curtiss, The energy of N2H2 and related compounds, J. Chem. Phys., 1991, 95, 4385–4388 CrossRef CAS
.
- T. A. Bazhenova and A. E. Shilov, Nitrogen fixation in solution, Coord. Chem. Rev., 1995, 144, 69–145 CrossRef CAS
.
- D. Singh, W. R. Buratto, J. F. Torres and L. J. Murray, Activation of Dinitrogen by Polynuclear Metal Complexes, Chem. Rev., 2020, 120, 5517–5581 CrossRef CAS PubMed
.
- S. G. Bratsch, Standard Electrode Potentials and Temperature Coefficients in Water at 298.15 K, J. Phys. Chem. Ref. Data, 1989, 18, 1–21 CrossRef CAS
.
- D. C. Rees, Great metalloclusters in enzymology, Annu. Rev. Biochem., 2002, 71, 221–246 CrossRef CAS PubMed
.
- O. Einsle and D. C. Rees, Structural Enzymology of Nitrogenase Enzymes, Chem. Rev., 2020, 120, 4969–5004 CrossRef CAS PubMed
.
- D. C. Rees, F. A. Tezcan, C. A. Haynes, M. Y. Walton, S. Andrade, O. Einsle and J. A. Howard, Structural basis of biological nitrogen fixation, Philos. Trans. R. Soc., A, 2005, 363, 971–984 CrossRef CAS PubMed
.
- Z.-Y. Yang, K. Danyal and L. C. Seefeldt, Mechanism of Mo-Dependent Nitrogenase, Methods Mol. Biol., 2011, 766, 9–29 CrossRef CAS PubMed
.
- T. Spatzal, M. Aksoyoglu, L. Zhang, S. L. A. Andrade, E. Schleicher, S. Weber, D. C. Rees and O. Einsle, Evidence for Interstitial Carbon in Nitrogenase FeMo Cofactor, Science, 2011, 334, 940 CrossRef CAS PubMed
.
- K. M. Lancaster, M. Roemelt, P. Ettenhuber, Y. Hu, M. W. Ribbe, F. Neese, U. Bergmann and S. DeBeer, X-ray Emission Spectroscopy Evidences a Central Carbon in the Nitrogenase Iron-Molybdenum Cofactor, Science, 2011, 334, 974–977 CrossRef CAS PubMed
.
- J. A. Wiig, Y. Hu, C. C. Lee and M. W. Ribbe, Radical SAM-Dependent Carbon Insertion into the Nitrogenase M-Cluster, Science, 2012, 337, 1672–1675 CrossRef CAS PubMed
.
- Y. Hu and M. Ribbe, A journey into the active center of nitrogenase, J. Biol. Inorg. Chem., 2014, 19, 731–736 CrossRef CAS PubMed
.
- C. H. Kim, W. E. Newton and D. R. Dean, Role of the MoFe protein alpha subunit histidine-195 residue in FeMo-cofactor binding and nitrogenase catalysis, Biochemistry, 1995, 34, 2798–2808 CrossRef CAS PubMed
.
- M. J. Dilworth, K. Fisher, C. H. Kim and W. E. Newton, Effects on substrate reduction of substitution of histidine-195 by glutamine in the alpha-subunit of the MoFe protein of Azotobacter vinelandii nitrogenase, Biochemistry, 1998, 37, 17495–17505 CrossRef CAS PubMed
.
- K. Fisher, M. J. Dilworth and W. E. Newton, Differential Effects on N2 Binding and Reduction, HD Formation, and Azide Reduction with a-195His- and a-191Gln-Substituted MoFe Proteins of Azotobacter Vinelandii Nitrogenase, Biochemistry, 2000, 39, 15570–15577 CrossRef CAS PubMed
.
- K. Fisher, M. J. Dilworth, C.-H. Kim and W. E. Newton, Azotobacter Vinelandii Nitrogenases Containing Altered MoFe Proteins with Substitutions in the FeMo-Cofactor Environment: Effects on the Catalyzed Reduction of Acetylene and Ethylene, Biochemistry, 2000, 39, 2970–2979 CrossRef CAS PubMed
.
- J. Christiansen, V. L. Cash, L. C. Seefeldt and D. R. Dean, Isolation and characterisation of an acetylene-resistant nitrogenase, J. Biol. Chem., 2000, 275, 11459–11464 CrossRef CAS PubMed
.
- J. Christiansen, L. C. Seefeldt and D. R. Dean, Competitive Substrate and Inhibitor Interactions at the Physiologically Relevant Active Site of Nitrogenase, J. Biol. Chem., 2000, 275, 36104–36107 CrossRef CAS PubMed
.
- P. M. C. Benton, J. Christiansen, D. R. Dean and L. C. Seefeldt, Stereospecificity of Acetylene Reduction Catalyzed by Nitrogenase, J. Am. Chem. Soc., 2001, 123, 1822–1827 CrossRef CAS PubMed
.
- P. M. C. Benton, M. Laryukhin, S. M. Mayer, B. M. Hoffman, D. R. Dean and L. C. Seefeldt, Localization of a Substrate Binding Site on the FeMo-Cofactor in Nitrogenase: Trapping Propargyl Alcohol with an R-70-Substituted MoFe Protein, Biochemistry, 2003, 42, 9102–9109 CrossRef CAS PubMed
.
- R. Y. Igarashi and L. C. Seefeldt, Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase., Crit. Rev. Biochem. Mol. Biol., 2003, 38, 351–384 CrossRef CAS PubMed
.
- B. M. Barney, R. Y. Igarashi, P. C. Dos Santos, D. R. Dean and L. C. Seefeldt, Substrate Interaction at an Iron-Sulfur Face of the FeMo-cofactor during Nitrogenase Catalysis, J. Biol. Chem., 2004, 279, 53621–53624 CrossRef CAS PubMed
.
- P. C. Dos Santos, R. Igarashi, H.-I. Lee, B. M. Hoffman, L. C. Seefeldt and D. R. Dean, Substrate Interactions with the Nitrogenase Active Site, Acc. Chem. Res., 2005, 38, 208–214 CrossRef CAS PubMed
.
- Z. Maskos, K. Fisher, M. Sorlie, W. E. Newton and B. J. Hales, Variant MoFe proteins of Azotobacter vinelandii: effects of carbon monoxide on electron paramagnetic resonance spectra generated during enzyme turnover, J. Biol. Inorg. Chem., 2005, 10, 394–406 CrossRef CAS PubMed
.
- K. Fisher, D. J. Lowe, P. Tavares, A. S. Pereira, B. H. Huynh, D. Edmondson and W. E. Newton, Conformations generated during turnover of the Azotobacter vinelandii nitrogenase MoFe protein and their relationship to physiological function, J. Inorg. Biochem., 2007, 101, 1649–1656 CrossRef CAS PubMed
.
- P. C. Dos Santos, S. M. Mayer, B. M. Barney, L. C. Seefeldt and D. R. Dean, Alkyne substrate interaction within the nitrogenase MoFe protein, J. Inorg. Biochem., 2007, 101, 1642–1648 CrossRef CAS PubMed
.
- L. C. Seefeldt, B. M. Hoffman and D. R. Dean, Mechanism of Mo-Dependent Nitrogenase, Annu. Rev. Biochem., 2009, 78, 701–722 CrossRef CAS PubMed
.
- B. M. Barney, H.-I. Lee, P. C. Dos Santos, B. M. Hoffman, D. R. Dean and L. C. Seefeldt, Breaking the N2 triple bond: insights into the nitrogenase mechanism, Dalton Trans., 2006, 2277–2284, 10.1039/b517633f
.
- R. Sarma, B. M. Barney, S. Keable, D. R. Dean, L. C. Seefeldt and J. W. Peters, Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an a-70Ile MoFe protein variant, J. Inorg. Biochem., 2010, 104, 385–389 CrossRef CAS PubMed
.
- S. M. Keable, J. Vertemara, O. A. Zadvornyy, B. J. Eilers, K. Danyal, A. J. Rasmussen, L. De Gioia, G. Zampella, L. C. Seefeldt and J. W. Peters, Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site, J. Inorg. Biochem., 2018, 180, 129–134 CrossRef CAS PubMed
.
- T. Spatzal, K. A. Perez, O. Einsle, J. B. Howard and D. C. Rees, Ligand binding to the FeMo-cofactor: Structures of CO-bound and reactivated nitrogenase, Science, 2014, 345, 1620–1623 CrossRef CAS PubMed
.
- T. Spatzal, K. A. Perez, J. B. Howard and D. C. Rees, Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor, eLife, 2015, 4, e11620 CrossRef PubMed
.
- D. Sippel, M. Rohde, J. Netzer, C. Trncik, J. Gies, K. Grunau, I. Djurdjevic, L. Decamps, S. L. A. Andrade and O. Einsle, A bound reaction intermediate sheds light on the mechanism of nitrogenase, Science, 2018, 359, 1484–1489 CrossRef CAS PubMed
.
- M. Rohde, K. Grunau and O. Einsle, CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution, Angew. Chem., Int. Ed., 2020, 59, 23626–23630 CrossRef CAS PubMed
.
- W. Kang, C. C. Lee, A. J. Jasniewski, M. W. Ribbe and Y. Hu, Structural evidence for a dynamic metallocofactor during N2 reduction by Mo-nitrogenase, Science, 2020, 368, 1381–1385 CrossRef CAS PubMed
.
- J. W. Peters, O. Einsle, D. R. Dean, S. DeBeer, B. M. Hoffman, P. L. Holland and L. C. Seefeldt, Comment on “Structural evidence for a dynamic metallocofactor during N2 reduction by Mo-nitrogenase”, Science, 2021, 371, eabe5481 CrossRef CAS PubMed
.
- W. Kang, C. C. Lee, A. J. Jasniewski, M. W. Ribbe and Y. Hu, Response to Comment on “Structural evidence for a dynamic metallocofactor during N2 reduction by Mo-nitrogenase”, Science, 2021, 371, eabe5856 CrossRef CAS PubMed
.
- T. M. Buscagan, K. A. Perez, A. O. Maggiolo, D. C. Rees and T. Spatzal, Structural Characterization of Two CO Molecules Bound to the Nitrogenase Active Site, Angew. Chem., Int. Ed., 2021, 60, 5704–5707 CrossRef CAS PubMed
.
- C. C. Lee, W. Kang, A. J. Jasniewski, M. T. Stiebritz, K. Tanifuji, M. W. Ribbe and Y. Hu, Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor, Nat. Catal., 2022, 5, 443–454 CrossRef CAS PubMed
.
- J. B. Varley, Y. Wang, K. Chan, F. Studt and J. K. Norskov, Mechanistic insights into nitrogen fixation by nitrogenase enzymes, Phys. Chem. Chem. Phys., 2015, 17, 29541–29547 RSC
.
- B. Benediktsson, A. T. Thorhallsson and R. Bjornsson, QM/MM calculations reveal a bridging hydroxo group in a vanadium nitrogenase crystal structure, Chem. Commun., 2018, 54, 7310–7313 RSC
.
- L. Cao, O. Caldararu and U. Ryde, Does the crystal structure of vanadium nitrogenase contain a reaction intermediate? Evidence from quantum refinement, J. Biol. Inorg. Chem., 2020, 25, 847–861 CrossRef CAS PubMed
.
- I. Dance, How feasible is the reversible S-dissociation mechanism for the activation of FeMo-co, the catalytic site of nitrogenase?, Dalton Trans., 2019, 48, 1251–1262 RSC
.
- P. E. M. Siegbahn, The mechanism for nitrogenase including all steps, Phys. Chem. Chem. Phys., 2019, 21, 15747–15759 RSC
.
- W. J. Wei and P. E. M. Siegbahn, A Mechanism for Nitrogenase Including Loss of a Sulfide, Chemistry, 2022, 28, e202103745 CrossRef CAS PubMed
.
- J. Bergmann, E. Oksanen and U. Ryde, Critical evaluation of a crystal structure of nitrogenase with bound N2 ligands, J. Biol. Inorg. Chem., 2021, 26, 341–353 CrossRef CAS PubMed
.
- H. Jiang and U. Ryde, Thermodynamically Favourable States in the Reaction of Nitrogenase without Dissociation of any Sulfide Ligand, Chem. – Eur. J., 2022, 28, e202103933 CrossRef CAS PubMed
.
- H. Jiang, O. K. G. Svensson, L. Cao and U. Ryde, Proton Transfer Pathways in Nitrogenase with and without Dissociated S2B, Angew. Chem., Int. Ed., 2022, 61, e202208544 CrossRef CAS PubMed
.
- J. Kastner and P. E. Blochl, Towards an Understanding of the Workings of Nitrogenase from DFT Calculations, ChemPhysChem, 2005, 6, 1–4 CrossRef PubMed
.
- J. Kastner, S. Hemmen and P. E. Blochl, Activation and protonation of dinitrogen at the FeMo cofactor of nitrogenase, J. Chem. Phys., 2005, 123, 074306 CrossRef PubMed
.
- J. Kastner and P. E. Blochl, Ammonia Production at the FeMo Cofactor of Nitrogenase: Results from Density Functional Theory, J. Am. Chem. Soc., 2007, 129, 2998–3006 CrossRef PubMed
.
- P. P. Hallmen and J. Kästner, N2 Binding to the FeMo-Cofactor of Nitrogenase, Z. Anorg. Allg. Chem., 2015, 641, 118–122 CrossRef CAS
.
- A. T. Thorhallsson, B. Benediktsson and R. Bjornsson, A model for dinitrogen binding in the E4 state of nitrogenase, Chem. Sci., 2019, 10, 11110–11124 RSC
.
- L. Cao and U. Ryde, What Is the Structure of the E4 Intermediate in Nitrogenase?, J. Chem. Theory Comput., 2020, 16, 1936–1952 CrossRef CAS PubMed
.
- I. Dance, Understanding the tethered unhooking and rehooking of S2B in the reaction domain of FeMo-co, the active site of nitrogenase, Dalton Trans., 2022, 51, 15538–15554 RSC
.
- I. Dance, Elucidating the Coordination Chemistry and Mechanism of Biological Nitrogen Fixation, Chem. – Asian J., 2007, 2, 936–946 CrossRef CAS PubMed
.
- I. Dance, Survey of the geometric and electronic structures of the key hydrogenated forms of FeMo-co, the active site of the enzyme nitrogenase: principles of the mechanistically significant coordination chemistry, Inorganics, 2019, 7, 8 CrossRef CAS
.
- I. Dance, Structures and reaction dynamics of N2 and H2 binding at FeMo-co, the active site of nitrogenase, Dalton Trans., 2021, 50, 18212–18237 RSC
.
- I. Dance, The pathway for serial proton supply to the active site of nitrogenase: enhanced density functional modeling of the Grotthuss mechanism, Dalton Trans., 2015, 44, 18167–18186 RSC
.
- I. Dance, What triggers the coupling of proton transfer and electron transfer at the active site of nitrogenase?, Dalton Trans., 2024, 53, 7996–8004 RSC
.
- I. Dance, Calculating the chemical mechanism of nitrogenase: new working hypotheses., Dalton Trans., 2022, 51, 12717–12728 RSC
.
- C. N. Morrison, J. A. Hoy, L. Zhang, O. Einsle and D. C. Rees, Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites, Biochemistry, 2015, 54, 2052–2060 CrossRef CAS PubMed
.
- D. Smith, K. Danyal, S. Raugei and L. C. Seefeldt, Substrate Channel in Nitrogenase Revealed by a Molecular Dynamics Approach, Biochemistry, 2014, 53, 2278–2285 CrossRef CAS PubMed
.
- I. Dance, The HD Reaction of Nitrogenase: a Detailed Mechanism, Chem. – Eur. J., 2022, 29, e202202502 CrossRef PubMed
.
- J. H. Guth and R. H. Burris, Inhibition of Nitrogenase-Catalyzed NH3 Formation by H2, Biochemistry, 1983, 22, 5111–5122 CrossRef CAS PubMed
.
- I. Dance, The binding of reducible N2 in the reaction domain of nitrogenase, Dalton Trans., 2023, 52, 2013–2026 RSC
.
-
R. N. F. Thorneley and D. J. Lowe, in Molybdenum enzymes, ed. T. G. Spiro, Wiley Interscience, New York, 1985, ch. 5, pp. 221–284 Search PubMed
.
- P. E. Wilson, A. C. Nyborg and G. D. Watt, Duplication and extension of the Thorneley and Lowe kinetic model for Klebsiella pneumoniae nitrogenase catalysis using a MATHEMATICA software platform, Biophys. Chem., 2001, 91, 281–304 CrossRef CAS PubMed
.
- D. F. Harris, A. Badalyan and L. C. Seefeldt, Mechanistic Insights into Nitrogenase FeMo-Cofactor Catalysis through a Steady-State Kinetic Model, Biochemistry, 2022, 61, 2131–2137 CrossRef CAS PubMed
.
- D. J. Lowe and R. N. F. Thorneley, The mechanism of klebsiella pneumoniae nitrogenase action. The determination of rate constants required for the simulation of the kinetics of N2 reduction and H2 evolution, Biochem. J., 1984, 224, 895–901 CrossRef CAS PubMed
.
- I. Dance, Unpublished results, 2024.
- L. Cao, O. Caldararu and U. Ryde, Protonation and Reduction of the FeMo Cluster in Nitrogenase Studied by Quantum Mechanics/Molecular Mechanics (QM/MM) Calculations, J. Chem. Theory Comput., 2018, 14, 6653–6678 CrossRef CAS PubMed
.
- C. Van Stappen, A. T. Thorhallsson, L. Decamps, R. Bjornsson and S. DeBeer, Resolving the structure of the E1 state of Mo nitrogenase through Mo and Fe K-edge EXAFS and QM/MM calculations, Chem. Sci., 2019, 10, 9807–9821 RSC
.
- I. Dance, Computational Investigations of the Chemical Mechanism of the Enzyme Nitrogenase, ChemBioChem, 2020, 21, 1671–1709 CrossRef CAS PubMed
.
- B. Benediktsson and R. Bjornsson, QM/MM Study of the Nitrogenase MoFe Protein Resting State: Broken-Symmetry States, Protonation States, and QM Region Convergence in the FeMoco Active Site, Inorg. Chem., 2017, 56, 13417–13429 CrossRef CAS PubMed
.
- L. Deng, H. Wang, C. H. Dapper, W. E. Newton, S. Shilov, S. Wang, S. P. Cramer and Z.-H. Zhou, Assignment of protonated R-homocitrate in extracted FeMo-cofactor of nitrogenase via vibrational circular dichroism spectroscopy, Commun. Chem., 2020, 3, 145 CrossRef CAS PubMed
.
- L. Cao, O. Caldararu and U. Ryde, Protonation States of Homocitrate and Nearby Residues in Nitrogenase Studied by Computational Methods and Quantum Refinement, J. Phys. Chem. B, 2017, 121, 8242–8262 CrossRef CAS PubMed
.
- T. Spatzal, J. Schlesier, E.-M. Burger, D. Sippel, L. Zhang, S. L. A. Andrade, D. C. Rees and O. Einsle, Nitrogenase FeMoco investigated by spatially resolved anomalous dispersion refinement, Nat. Commun., 2016, 7, 10902 CrossRef CAS PubMed
.
- R. Bjornsson, F. Neese and S. DeBeer, Revisiting the Mössbauer Isomer Shifts of the FeMoco Cluster of Nitrogenase and the Cofactor Charge, Inorg. Chem., 2017, 56, 1470–1477 CrossRef CAS PubMed
.
- B. Delley, An all-electron numerical method for solving the local density functional for polyatomic molecules, J. Chem. Phys., 1990, 92, 508–517 CrossRef CAS
.
-
B. Delley, in Modern density functional theory: a tool for chemistry, ed. J. M. Seminario and P. Politzer, Elsevier, Amsterdam, 1995, vol. 2, pp. 221–254 Search PubMed
.
- J. Baker, A. Kessi and B. Delley, The generation and use of delocalized internal coordinates in geometry optimization, J. Chem. Phys., 1996, 105, 192–212 CrossRef CAS
.
- B. Delley, From molecules to solids with the DMol3 approach, J. Chem. Phys., 2000, 113, 7756–7764 CrossRef CAS
.
- J. Andzelm, R. D. King-Smith and G. Fitzgerald, Geometry optimization of solids using delocalized internal coordinates, Chem. Phys. Lett., 2001, 335, 321–326 CrossRef CAS
.
- T. Todorova and B. Delley, Molecular Crystals: A Test System for Weak Bonding, J. Phys. Chem. C, 2010, 114, 20523–20530 CrossRef CAS
.
- J. P. Perdew, K. Burke and M. Ernzerhof, Generalized Gradient Approximation Made Simple, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed
.
- I. Dance, Evaluations of the Accuracies of DMol3 Density Functionals for Calculations of Experimental Binding Enthalpies of N2, CO, H2, C2H2 at Catalytic Metal Sites, Mol. Simul., 2018, 44, 568–581 CrossRef CAS
.
- J. Andzelm, C. Kolmel and A. Klamt, Incorporation of solvent effects into density functional calculations of molecular energies and geometries, J. Chem. Phys., 1995, 103, 9312–9320 CrossRef CAS
.
- A. Klamt, V. Jonas, T. Burger and J. C. W. Lohrenz, Refinement and Parametrization of COSMO-RS, J. Phys. Chem. A, 1998, 102, 5074–5085 CrossRef CAS
.
- B. Delley, The conductor-like screening model for polymers and surfaces, Mol. Simul., 2006, 32, 117–123 CrossRef CAS
.
- L. Cao and U. Ryde, Influence of the protein and DFT method on the broken-symmetry and spin states in nitrogenase, Int. J. Quantum Chem., 2018, 118, e25627 CrossRef
.
- S. Raugei, L. C. Seefeldt and B. M. Hoffman, Critical computational analysis illuminates the reductive-elimination mechanism that activates nitrogenase for N2 reduction, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E10521–E10530 CrossRef CAS PubMed
.
- A. T. Thorhallsson and R. Bjornsson, The E2 state of FeMoco: Hydride Formation versus Fe Reduction and a Mechanism for H2 Evolution, Chem. – Eur. J., 2021, 27, 16788–16800 CrossRef CAS PubMed
.
- H. Jiang, O. K. G. Svensson and U. Ryde, QM/MM Study of Partial Dissociation of S2B for the E2 Intermediate of Nitrogenase, Inorg. Chem., 2022, 61, 18067–18076 CrossRef CAS PubMed
.
- Y. Pang and R. Bjornsson, Understanding the Electronic Structure Basis for N2 Binding to FeMoco: A Systematic Quantum Mechanics/Molecular Mechanics Investigation, Inorg. Chem., 2023, 62, 5357–5375 CrossRef CAS PubMed
.
- Y. Pang and R. Bjornsson, The E3 state of FeMoco: one hydride, two hydrides or dihydrogen?, Phys. Chem. Chem. Phys., 2023, 25, 21020–21036 RSC
.
- H. Jiang and U. Ryde, N2 binding to the E0–E4 states of nitrogenase, Dalton Trans., 2023, 52, 9104–9120 RSC
.
- I. Dance, A pragmatic method for location of transition states and calculation of reaction paths., Mol. Simul., 2008, 34, 923–929 CrossRef CAS
.
- I. Dance, A pragmatic method for location of transition states and calculation of reaction paths: erratum, Mol. Simul., 2011, 37, 257 CrossRef CAS
.
- A. Kohen and J. P. Klinman, Enzyme Catalysis: Beyond Classical Paradigms., Acc. Chem. Res., 1998, 31, 397–404 CrossRef CAS
.
- D. Lukoyanov, Z.-Y. Yang, N. Khadka, D. R. Dean, L. C. Seefeldt and B. M. Hoffman, Identification of a Key Catalytic Intermediate Demonstrates That Nitrogenase Is Activated by the Reversible Exchange of N2 for H2, J. Am. Chem. Soc., 2015, 137, 3610–3615 CrossRef CAS PubMed
.
- D. Lukoyanov, N. Khadka, Z.-Y. Yang, D. R. Dean, L. C. Seefeldt and B. M. Hoffman, Reductive Elimination of H2 Activates Nitrogenase to Reduce the N
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N Triple Bond: Characterization of the E4(4H) Janus Intermediate in Wild-Type Enzyme, J. Am. Chem. Soc., 2016, 138, 10674–10683 CrossRef CAS PubMed
N Triple Bond: Characterization of the E4(4H) Janus Intermediate in Wild-Type Enzyme, J. Am. Chem. Soc., 2016, 138, 10674–10683 CrossRef CAS PubMed .
- D. Lukoyanov, N. Khadka, Z.-Y. Yang, D. R. Dean, L. C. Seefeldt and B. M. Hoffman, Reversible Photoinduced Reductive Elimination of H2 from the Nitrogenase Dihydride State, the E4(4H) Janus Intermediate, J. Am. Chem. Soc., 2016, 138, 1320–1327 CrossRef CAS PubMed
.
- I. Dance, A molecular pathway for the egress of ammonia produced by nitrogenase, Sci. Rep., 2013, 3, 3237 CrossRef PubMed
.
- J. L. Crossland and D. R. Tyler, Iron–dinitrogen coordination chemistry: Dinitrogen activation and reactivity, Coord. Chem. Rev., 2010, 254, 1883–1894 CrossRef CAS
.
- J. Rittle and J. C. Peters, An Fe-N2 Complex That Generates Hydrazine and Ammonia via Fe
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NNH2: Demonstrating a Hybrid Distal-to-Alternating Pathway for N2 Reduction, J. Am. Chem. Soc., 2016, 138, 4243–4248, DOI:10.1021/jacs.6b01230
NNH2: Demonstrating a Hybrid Distal-to-Alternating Pathway for N2 Reduction, J. Am. Chem. Soc., 2016, 138, 4243–4248, DOI:10.1021/jacs.6b01230 .
- R. J. Burford and M. D. Fryzuk, Examining the relationship between coordination mode and reactivity of dinitrogen, Nat. Rev. Chem., 2017, 1, 0026 CrossRef CAS
.
- T. M. Buscagan and D. C. Rees, Rethinking the Nitrogenase Mechanism: Activating the Active Site, Joule, 2019, 3, 2662–2678 CrossRef CAS PubMed
.
- Y. Nishibayashi, Development of catalytic nitrogen fixation using transition metal–dinitrogen complexes under mild reaction conditions, Dalton Trans., 2018, 47, 11290–11297 RSC
.
- I. Dance, Activation of N2, the Enzymatic Way, Z. Anorg. Allg. Chem., 2015, 641, 91–99 CrossRef CAS
.
- B. M. Barney, J. McClead, D. Lukoyanov, M. Laryukhin, T. C. Yang, D. R. Dean, B. M. Hoffman and L. C. Seefeldt, Diazene (HN=NH) Is a Substrate for Nitrogenase: Insights into the Pathway of N2 Reduction, Biochemistry, 2007, 46, 6784–6794 CrossRef CAS PubMed
.
- L. Cao and U. Ryde, N2H2 binding to the nitrogenase FeMo cluster studied by QM/MM methods, J. Biol. Inorg. Chem., 2020, 25, 521–540 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Description of the protein model and the structural constraints used, including movement of Val70 and the contiguous chain, a description of the density functional procedures and their validation, a description of procedures for determination of transition states and reaction trajectories, and atomic coordinates for reactants, transition states and products. See DOI: https://doi.org/10.1039/d4dt01866d |
| This journal is © The Royal Society of Chemistry 2024 |