Open Access Article
Open Access ArticleOptimising Shifted Excitation Raman Difference Spectroscopy (SERDS) for application in highly fluorescent biological samples, using fibre optic probes†
H.
Sheridan
a,
A. P.
Dudgeon
 abc,
J. C. C.
Day
c,
C.
Kendall
ab,
C.
Hall
d and
N.
Stone
abc,
J. C. C.
Day
c,
C.
Kendall
ab,
C.
Hall
d and
N.
Stone
 *ab
*ab
aBiomedical Physics, Department of Physics and Astronomy, University of Exeter, Exeter, EX4 4QL, UK
bBiophotonics Research Unit, Gloucestershire Hospitals NHS Foundation Trust, Gloucester, GL1 3NN, UK
cInterface Analysis Centre, HH Wills Physics Laboratory, Tyndall Avenue, University of Bristol, BS8 1TL, UK
dDepartment of Otolaryngology and Head & Neck Surgery, Gloucestershire Hospitals NHS Foundation, Trust, GL53 7AN, UK
First published on 19th November 2024
Abstract
Fibre optic probe based Raman spectroscopy can deliver in vivo molecular compositional analysis of a range of diseases. However, some biological tissues exhibit high levels of fluorescence which limit the utility of the technique, particularly when the fluorescence induces CCD etaloning, which can be particulalry hard to remove in subsequent analysis. Furthermore, use of fibre probes can result in silica signals superimposed on the biological Raman signals. Shifted excitation Raman difference spectroscopy (SERDS) utilises a small seperation in excitation wavelengths to remove signals from fluorescence, room lights, optical components and etaloning contributions, while retaining chemical signals from the sample. In this study, we sought to measure the optimum SERDS spectra enabling reconstruction of a range a narrow and broad peaks found in biological samples. A original wavelength of 830 nm was utilised with 7 different shifts between 0.4 and 3.9 nm to determine which gave the best performance. This range roughly corresponds to the typical range of peak widths within biological Raman spectra at 830 nm excitation; 0.41 – 3.25 nm or 6 – 47 cm−1. An wavelength shift of 2.4 nm was identified as optimal. Finally, a fibre optic Raman probe was used to measure 2 human lymph nodes ex vivo to demonstrate the feasibility of the approach with real-world examples.
Introduction
Raman spectroscopy is a label free, non-invasive technique that provides highly specific biochemical analysis of samples.1–4 Its ability to probe the vibrational modes of the chemical bonds within a sample has led to widespread investigation within the area of disease diagnosis.1,5,6 Chemometric models based on Raman data can achieve sensitivities and specificities up to or above 90%.7–11 Thus, Raman spectroscopy represents a tool with the ability to outperform classical disease diagnosis methods; histopathology of cancerous tissues is a process prone to both inter- and intra- observer disagreements, particularly for intermediate disease states.12–18In order to facilitate in vivo diagnosis, anatomical access requirements typically require the use of fibre optic Raman probes, which are amenable to incorporation into the existing diagnostic pathway.19–21 As such, Raman probes are being developed for a wide variety of diseases and locations, including cancers of the bladder,22,23 GI tract,24–26 cervix,27,28 breast,29,30 brain7,21 and lymphatic system.31,32 The development of such probes has greatly improved the clinical applicability of the Raman technique,19,20,33 however the use of silica fibres typically used within these designs adds to the unwanted background contributions.19,20,24,34,35
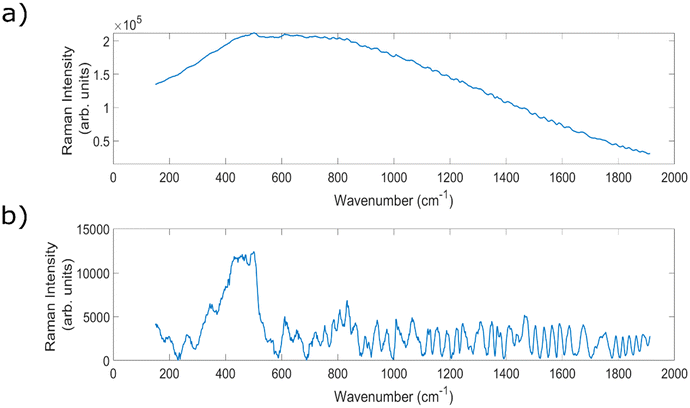
Within the field, a common barrier to this in vivo implementation can be the presence of strong auto fluorescence signals when analysing biological samples, threatening to overwhelm the chemically-specific, diagnostically important peaks.5,36 An example of a spectrum with high levels of fluorescence, and the resulting baselined spectrum, can be seen in Fig. 1. Raman scattering is a relatively weak effect, occurring for approximately one in every 106 incident photons, meaning that it can be easily overwhelmed by the much stronger fluorescence signals.36 The strength of these fluorescence signals varies across different sample types;37,38 lymph nodes,39,40 prostate41 and breast tissue40 have all demonstrated high levels of fluorescence.
Firstly, the fluorescence signals are accompanied by an increase in the shot noise in the spectrum (±√N), where N is the measured photoelectron count in each pixel of the CCD. This can overwhelm Raman signals, making simple background subtraction ineffective. Moreover, the background signals produced by fluorescence can exacerbate the presence of an optical effect known as etaloning, which serves to further obfuscate true Raman signals.20 Etaloning is a process wherein photons in the photosensitive region of the CCD detector are reflected at each region's boundary, and constructively and destructively interfere with incoming photons to create a ‘ringing’ pattern within the spectrum, that is wavelength dependant.1,42–45 These effects can combine to reduce the efficacy of developed in vivo Raman diagnostic tools.
There are two main types of method for the reduction or suppression of these unwanted fluorescence signals; hardware and mathematical approaches. Hardware approaches range from modifying the coatings or changing the CCD thickness, shifting the excitation wavelength towards the Mid-IR region, to time gated and frequency domain techniques.36,46 Each of these techniques has associated drawbacks. A shift towards Mid-IR excitation wavelengths reduces the excitation of the electronic transitions associated with auto-fluorescence, however it also reduces the intensity of Raman signals thanks to the λ−4 dependence of Raman scattering.2,33,37 CCD quantum efficiency also decreases. Time gating approaches can be applied, whereby almost instantaneous Raman scattering can be separated from delayed (ps) fluorescence emissions. However, these approaches can suffer from limited clinical applicability because of the high cost of the required specialist equipment,47 and also are not successful in applications wherein the fluorescence lifetime is comparable to the laser pulse length or the time resolution of the methodology.36,46 Frequency domain methods also require a complicated experimental setup, which could create complications for clinical translation by increasing the overall footprint of the setup and reducing portability, or by making standardisation of the technology more difficult.48 Moreover, these techniques are less effective in reducing the impact of fluorescence when measuring samples with low Raman intensity, such as for measurement of biologicals.36,46
Mathematical approaches involve using different algorithms to computationally remove the broadband background signals after data acquisition.20,49 Whilst there are number of methods that can achieve this at no extra experimental cost (for example: polynomial fitting,20,49–51 least squares methods,49,52 and extended multiplicative scatter correction (EMSC)50–52), if the background signals have overwhelmed the Raman signals, the application of these techniques may introduce significant distortions to the spectral information which can reduce the diagnostic potential. Each of these techniques has to be tuned to each specific spectra analysed, which is time consuming, and often struggles with structured spectral backgrounds, such as those from lights, optics, and CCD etaloning contributions. For highly fluorescent samples such as liver, fluorescence levels are so high that separating the Raman signals from the background is not usually possible.20,49
Shifted excitation Raman difference spectroscopy (SERDS) is an alternative technique to remove fluorescence and other unwanted spectral features. The underlying theory of the technique is that a small shift in excitation wavelength corresponds to a small shift in the Raman peak bands, and no change to the broadband fluorescence contributions. Thus, by obtaining two spectra at slightly shifted excitation wavelengths, and then subtracting one from the other, one can theoretically obtain a difference spectrum which contains the chemically specific information free from the fluorescence contributions. This difference spectrum can then be integrated as a function of the wavelength shift utilised in order to obtain a ‘pure’ Raman spectrum, free from undesirable background signals.36,49–52 Another benefit of the technique is its ability to simultaneously remove etaloning signals from the obtained spectra.50,52–54
Since being first reported in 1992,55 the SERDS technique has been investigated in a variety of fields, including food inspection,56 security,57 agricultural58,59 and biological analysis,49,50,59 and conservation.60,61 It has also been applied to a large variety of different samples, ranging from ex vivo human tissues,62 bacteria,63 fungi,64 heritage materials60,65 to polymers.66 In fact, the technique has even recently been combined with a fibre optic Raman probe design for in vivo assessment of nasopharyngeal tissue, representing the first use of endoscopic SERDS within a human cavity.67 Previous studies have demonstrated the ability of the technique to remove strong fluorescent background contributions even within spectra where the background intensity outweighs the Raman signal in a ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.52
1.52
Traditionally, the ideal wavelength gap between excitation wavelengths has been defined as half of the spectral bandwidth of a typical Raman peak within the spectra. However, this value can vary significantly within spectra of biological tissues, both between and within different sample types, and as such this criterion does not necessarily represent the optimal choice.52,62 A 2017 study comparing shifts of 0.5, 1.0, 1.5 and 2.0 nm with 785 nm excitation determined experimentally that the 2 nm gap provided the highest quality SERDS spectra. Furthermore, within simulations completed in the same work, the theoretical optimal shift was identified as 10 nm for protein samples and 7 nm for lipid samples. The authors reported that shifts larger than 2 nm were not possible to analyse, owing to the filtration constraints of the optical setup.49
A 2015 study using 785 nm excitation to analyse human oral tissue compared shifts of 0.1, 0.3, 0.4, 0.5, 0.6, 0.9 and 1.0 nm, and determined that 1.0 nm gave the best results.49 A 2020 study measuring pollen samples compared shifts of 1.0 and 2.0 nm, and found that a 2.0 nm conferred optimal performance.50 Given that Raman peaks can range in width from 6–47 cm−1 (which corresponds to 0.41–3.25 nm at 830 nm excitation),68–70 even if the half width criteria is accepted, this leads to a wide range of possible ideal wavelength shifts. It is therefore valuable to compare as wide a range of excitation shifts as possible within the experimental setup, in order to determine the most effective shift to remove fluorescence and etaloning contributions, whilst maximising spectral quality. In addition, the use of the SERDS technique can facilitate a dramatic reduction in the silica luminescent background signals generated from the optical fibres.
In this study 7 different excitation shifts, ranging from 0.4 to 3.9 nm, were studied in a variety of biological samples in order to determine which shift gave the optimal SERDS spectra. To the best of the authors’ knowledge, this represents the largest wavelength range tested in an experimental comparison of SERDS excitation gaps. After selection of the optimal experimental parameters, proof of concept measurements on two ex vivo lymph node samples were performed.
Experimental methods
SERDS setup
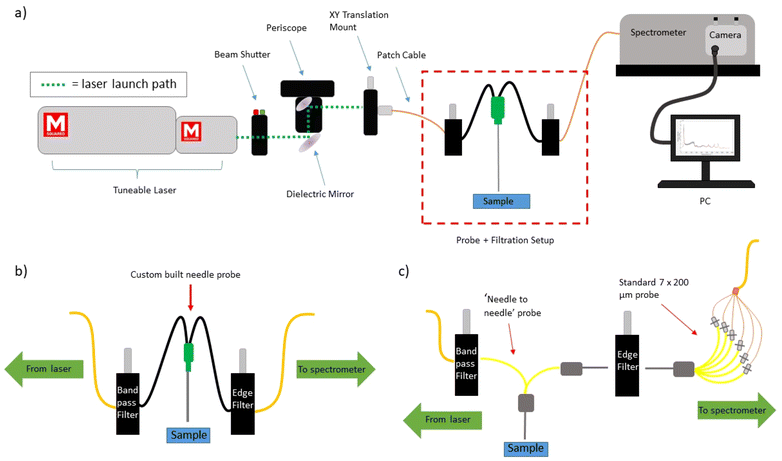
A tuneable Ti:Sapphire laser (M2 Lasers, SolsTIS 5000, Glasgow, UK) was used to achieve each of the wavelengths studied in this work (830.0 nm–833.9 nm). The laser light was launched into the ‘delivery optics’ and then into a 200 μm, 0.22 NA patch cable. The ‘delivery optics’ consisted of a laser safety beam shutter (Lasermet, LS-10, Dorset, UK), followed by a complete periscope assembly (Thorlabs, RS99/M, New Jersey, USA) with attached broadband dielectric mirror (Thorlabs, BB1-E03, New Jersey, USA). Subsequent to reflection by the periscope mirror system, the laser light was focused onto a FC/PC adapter (Thorlabs, SM1FC, New Jersey, USA) contained within a XY translation mount (Thorlabs, CXY1A, New Jersey, USA), to which the 200 μm, 0.22 NA patch cable was attached. The laser light was then filtered by an 830 nm band pass filter (Thorlabs L-830-10 #8835), housed within an in-line multi-mode fibre optical filter mount (Thorlabs, FOFMF/M, New Jersey, USA). A schematic of the experimental setup can be seen in Fig. 2a.Within this work, two different Raman probe designs were used and will be detailed in the following subsection. Both probe designs utilised a long pass edge filter (Semrock, LP02-830RU-25, New York, USA) within the same type of in-line optical mount to filter the collected Raman signals. This light was then delivered to the entrance port of a Holospec 1.8i Kaiser spectrometer (Kaiser Optical Systems, HOLOSPEC-F/1.8-NIR, Ann Arbor, USA). The internal slit size within the spectrometer (Kaiser Optical Systems, SLIT-50, Ann Arbor, USA) was 50 μm, and an Andor iDus 420 was used as the CCD (Andor – Oxford Instruments, DU420A-BR-DD, Abingdon, UK). The spectral resolution of the system was ∼3.5 cm−1.
The system was wavelength calibrated using a NeAr source (Renishaw, Wootton-under-Edge, UK). Aspirin measurements were also taken at each of the excitation wavelengths measured in order to accurately monitor the shift in the Raman peaks. Other standards (green glass, stainless steel, PTFE) were also measured to monitor system performance.
For all SERDS measurements, the CCD readout rate was 33 kHz. The excitation wavelengths measured were: 830.0 nm and seven shifted wavelengths listed for ease of description in Table 1. For each SERDS spectrum, one shifted wavelength spectra was subtracted from an 830.0 nm excitation spectrum of the same acquisition time. During the calculation of these SERDS difference spectra, the k factor method (described by Korinth, F. et al.51) was used to multiply the shifted excitation spectrum by a factor ‘−k’, prior to subtracting it from the 830.0 nm spectra. The resulting difference spectrum was then ‘-zero centered’ (see Gebrekidan et al.49) by the application of a 1D median filter with a frame length of 100.
| Wavelength shift number | Shifted wavelength (nm) |
|---|---|
| 1 | 830.4 |
| 2 | 830.9 |
| 3 | 831.6 |
| 4 | 832.0 |
| 5 | 832.4 |
| 6 | 833.3 |
| 7 | 833.9 |
For each measurement position on the sample surface, one 830.0 nm excitation spectrum was taken with an acquisition time equal to the combined total of the SERDS spectrum, all taken in the same location (to ensure a fair comparison of performance).
Raman probe designs
For all of the animal tissue measurements within this work, a simple 1 in 1 out fibre optic ‘Raman needle probe #1′ was used; schematically described within (Fig. 2b). It was created in house and consisted of a 20G thinwall hypodermic needle (Coopers Needle Works, Birmingham, UK) of 50 mm length with 2 × 200 μm, 0.22 NA fibres (Thorlabs, FG200LEA, New Jersey, USA) affixed inside with optically clear, fast cure, medical grade adhesive (Intertronics, OPT5006-1-50G, Kidlington, UK). One fibre was used for delivery and one for collection. The collection fibre connected directly to the inline filter housing using an FC connector, from where a 200 μm, 0.22 NA patch cable delivered the laser light to the spectrometer. The tip of the probe was polished flat, and the power at the probe tip for all measurements was ∼150–160 mW (provided by a pump laser power of 6.9 W).For the lymph node measurements, a more complex probe design was used; ‘Raman needle probe #2′, demonstrated schematically in (Fig. 2c). This setup was used as it conferred higher overall signal levels than Raman needle probe #1, and thus a reduction in acquisition time. The design consists of 1 × 200 μm, 0.22 NA laser delivery fibre (Thorlabs, FG200LEA, New Jersey, USA), surrounded by 6 × 200 μm, 0.22 NA collection fibres, all contained within the bore of a 19G hypodermic needle (Coopers Needle Works, Standard Size 19G Stainless Steel Tubing, Birmingham, UK). This configuration is termed ‘6 around 1′, as the laser delivery fibre is the central fibre, and the 6 collection fibres form a ring around it. An FC connector (Thorlabs, 30230C1, New Jersey, USA) connects the laser delivery fibre to the bandpass filter via the previously described in-line multi-mode optical filter mount. The 6 collection fibres are terminated within another 19G needle core; edge filtration is achieved using the previously described optical mount along with coupling to 7 × 200 μm collection fibres within a 19G needle core. Post mountable V groove clamps (Thorlabs, VC1, New Jersey, USA) are used to mount and perform a simple alignment of the two needle cores. The 7 × 200 μm collection fibres then deliver the collected signals to the spectrometer via a custom-built FC connector to SMA linear bundle (Thorlabs, FG105LCA-FBUNDLE, New Jersey, USA). All fibres were affixed in place within the needle cores using optically clear, fast cure, medical grade adhesive (Intertronics, OTP5006-1-50G, Kidlington, UK). The tips of each of the needles were polished flat, and the power at the probe tip for all measurements was ∼150–160 mW.
Raman microscope measurements
A Raman microscope (Renishaw, inVia confocal Raman microscope, BR010, Wotton-under-Edge, UK) was used to measure samples using standard Raman spectroscopy with 830 nm excitation. The system was calibrated using measurements of silicon, an internal neon argon emission source, and a fluorescent green glass sample. The laser power was set to 100% (around 130 mW at the sample), a 50× objective was used and the grating was 1200 l mm−1. Acquisition time was 3 × 1 s.The samples measured were thick cut smoked porcine muscle, and chicken breast (Tesco PLC, Welwyn Garden City, UK), cut into ∼2 cm × 2 cm pieces. Samples were removed from storage at −80 °C and allowed to defrost at room temperature for ∼30 minutes, before being placed onto a custom stainless steel slide (Grade 304 super mirror stainless steel, Ulbrich, UNS S30400, Connecticut, USA, with dimensions of 75 mm × 24 mm × 0.9 mm) for measurement.
SERDS measurements
Samples were thawed at room temperature for ∼15 minutes prior to measurements. Samples were placed on a CaF2 substrate for Raman spectral measurements and then snap frozen using liquid nitrogen before being returned to the freezer. Sample 1 was confirmed via histology report to be high-grade non-Hodgkin lymphoma, and sample 2 was a reactive lymph node (non-cancerous inflammatory). For each lymph node sample, there were 7 measurement positions per sample, with 5 spectra taken at each wavelength at each position. All spectra were obtained using Raman needle probe #2. The acquisition time for the individual spectra used to create the difference spectra was 10 s, and for the standard Raman measurement the acquisition time was 20 s.
Data processing
All data analysis was carried out in MATLAB R2018a. Some spectra were excluded upon visual inspection as a result of the presence of cosmic rays (10 out of 240 spectra). Where baselining was used, an asymmetric least squares fit was utilised. All principal component analysis (PCA) was carried out using the inbuilt Matlab function ‘pca’, which includes mean centring as a default pre-processing step, followed up with use of a student t-test to determine the most statistically significant PCs. Throughout the manuscript, where SERDS data is referred to as ‘normalised’, this refers to the effects of the k factor and mean centering processes.Within this work, signal to noise ratio (SNR) and signal to background (SBR) metrics were routinely calculated. For the SNR measurements, the peak of interest was isolated from the spectrum, baselined and then peak intensity calculated. For the SERDS spectra, the height of the peak was calculated as the height above the zero line. This peak intensity value was then ratioed with the standard deviation of 5 pixels within a flat region of the spectrum/difference spectrum. SBR was calculated by ratioing the peak intensity with the average absolute value of 30 pixels of the broad silica background region, defined as the area preceding the sharp silica peaks at ∼426 and 485 cm−1.
Results and discussion
Shift comparisons
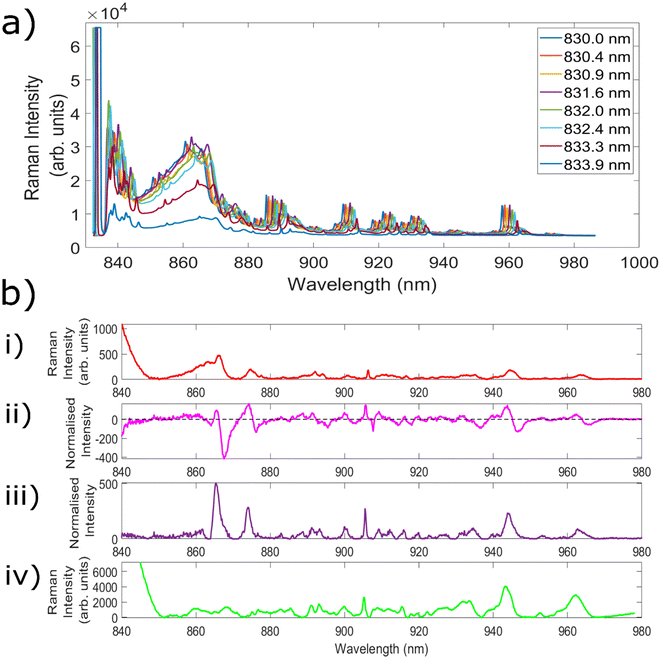
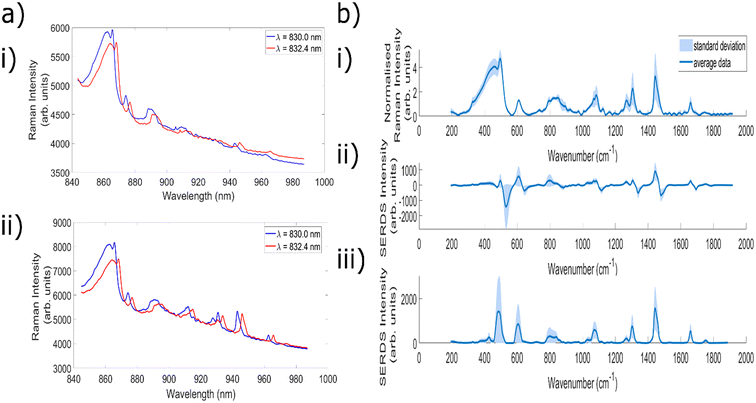
Fig. 3a demonstrates measurements of Aspirin using the 8 different excitation wavelengths utilised to create the 7 different wavelength shifts. The separation between each of the wavelengths can most clearly be seen for the series of peaks centred at ∼960 nm. From the excitation wavelength of 831.6 nm there is a slight decrease in peak intensity with each increase in wavelength, leading to a significantly reduced intensity for the peak obtained with 833.9 nm excitation. This is because the laser line filters used within this setup are optimised for 830 nm excitation, and as such their transmission decreases outside of the optimum ‘transmission window’, before eventually being blocked altogether. Nevertheless, the ability to retain the same filters as utilised within a standard Raman setup simplifies both the instrumental requirements and the cost associated with the setup. As a result, each of the wavelengths from 830.4 nm onwards was combined with standard 830.0 nm excitation in order to create 7 different SERDS wavelength shifts.The next experiments measured both porcine muscle and chicken samples with the 7 different wavelength shifts, before comparing the resulting SERDS difference and reconstructed spectra with the equivalent ‘standard’ Raman spectra. Samples of porcine tissue and chicken were chosen owing to their similar optical and mechanical properties as compared with human tissue.71 Within the standard Raman measurements taken with the fibre optic probes, an acquisition time was used equivalent to the combined acquisition time of each of the individual spectra used to create the difference spectrum. This is because the noise of each individual spectrum is additive within the resulting difference spectrum, and therefore this represents the most truthful comparison to typical Raman spectra.52 Given that porcine and chicken muscle samples are not associated with high levels of fluorescence, the aim of these initial tests was to evaluate the efficacy of the SERDS technique in retrieving chemical signals from the sample of interest, as well remove silica background signals.
Fig. 3b(i) shows the average spectrum of porcine muscle taken using the Raman needle probe #1. Fig. 3b(ii) shows the Raman difference spectrum of porcine muscle obtained using excitation wavelengths of 830.0 nm and 831.6 nm. Fig. 3b(iii) shows the corresponding reconstructed SERDS spectrum, and Fig. 3b(iv) demonstrates the average porcine muscle spectrum taken using a Raman microscope. The peak at 905.5 nm corresponds to a Raman shift of 1004.6 cm−1 and is identified as the phenylalanine peak of porcine muscle tissue. The peaks at 915.5 nm (1125.2 cm−1), 943.8 nm (1452.7 cm−1) and 962.2 nm (1655.3 cm−1) correspond to the C–C skeletal stretch, CH3 anti-symmetric stretch and CH2–CH bend and amide I, alpha-helical structures of porcine muscle tissue, respectively.72 Each of the different types of spectra shown within this figure demonstrate good agreement with the porcine peaks identified within the literature, confirming the ability of the SERDS technique to retain chemical information from the sample.
The average spectrum taken using needle probe #1 shows both silica background contributions (below ∼880 nm) and signals originating from the porcine sample (seen particularly clearly above 900 nm). The SERDS difference spectrum contains the same spectral signatures as the standard Raman needle probe #1 spectrum, but the peaks are replaced with first derivative line shapes. This is seen most clearly in the peak at ∼905 nm. With regards to the silica contributions, the contributions between ∼850–865 nm are significantly reduced within the difference spectrum, although specific Raman peaks corresponding to the silica fibres (865 nm, 874 nm) are not removed in the same way as the broad background luminescent shape is.
The reconstructed SERDS spectrum appears to demonstrate the same chemical peaks seen within the standard Raman needle probe #1 spectrum, with a significantly reduced broad background region (∼850–865 nm) and an increase to the relative sample contributions. This can be seen as an increase to the intensity of the peaks originating from the sample itself. Furthermore, a comparison between the SERDS reconstruction and the spectrum taken using the Raman microscope (a spectrum free of the silica contributions that result when measuring with fibre optic probes) indicates strong agreement between the two spectra. The silica peaks (865 nm, 874 nm) are still present within the reconstructed spectra, however similarities between the spectral signatures above 880 nm indicate that the SERDS technique maintains important chemical signals originating from within the sample.
Given that the technique maintains the spectral contributions from the sample whilst also significantly reducing the silica background, each of the 7 wavelength shifts identified within Fig. 3a were compared to determine which combination of excitation wavelengths led to the largest improvement to data quality, and most effective reduction in fluorescence and etaloning levels.
Highly fluorescent samples
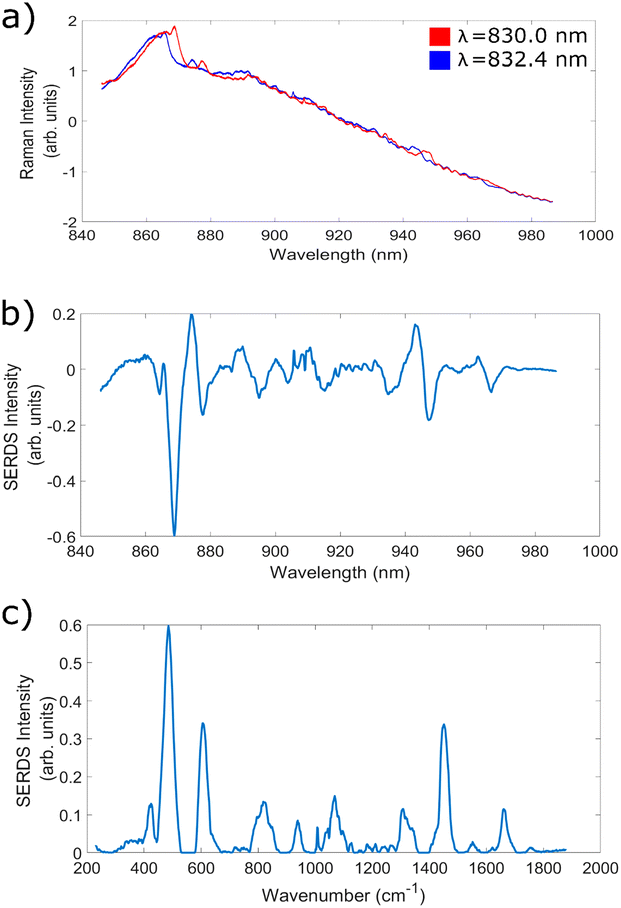
In the next set of experiments, bovine cheek and porcine liver samples were utilised to facilitate measurements with high levels of fluorescence and investigate the ability of the SERDS technique to remove these contributions, across each of the 7 wavelength shifts identified within the previous subsection. Fig. 4a shows an example of two highly fluorescent spectra taken at wavelengths corresponding to wavelength shift 5 (830.0 and 832.4 nm), Fig. 4b shows the corresponding SERDS difference spectra, and Fig. 4c shows the SERDS reconstruction. The baselined version of the 830 nm excitation spectra in Fig. 4a can be seen in ESI Fig. 1.† It is clear to see that the etaloning contributions within the raw spectra are successfully removed within both types of SERDS spectra. It can also be clearly seen from ESI Fig. 2† that simply digitally shifting the original bovine cheek spectra by the same amount of nm as achieved experimentally does not remove the etaloning contributions to the resulting difference spectrum. Furthermore, this technique acts to obfuscate the Raman peaks within the resulting difference spectrum.This data demonstrates the ability of the SERDS technique to successfully remove both general fluorescence background and etaloning contributions from spectra, both of which are important when measuring highly fluorescent tissue. It is also clear from this data that the resulting difference and reconstructed spectra are able to retain chemically relevant peaks originating from within the sample; the peaks at 1004 cm−1 (phenylalanine31,72), 1449 cm−1 (CH2 bend20,73) and 1661 cm−1 (amide 120,72,74) are commonly associated with biological samples. Therefore, for the next stage of analysis, each of the different wavelength shifts were compared to see which combination gave the greatest reduction in fluorescence and etaloning contributions and best overall data quality.
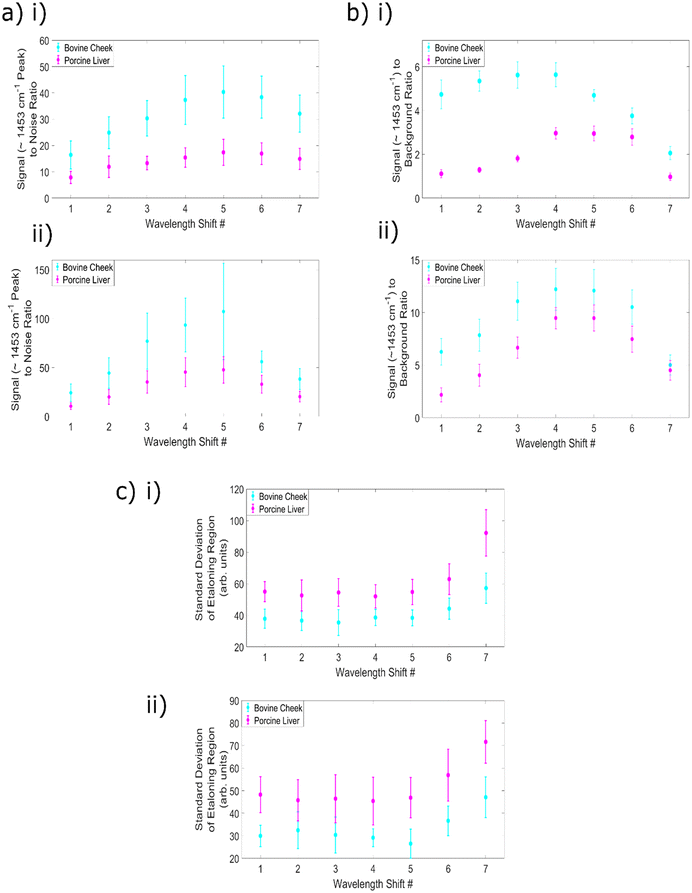
An evaluation of the effect of changing wavelength shift number on the achievable signal to noise ratio is an important aspect of any assessment of the overall data quality, especially given that the high levels of fluorescence within these samples dramatically increases the overall count number and therefore the noise levels within the spectra. The results of this analysis can be seen within Fig. 5a(i) and (ii) with subplot (i) containing the difference spectra data and (ii) the reconstructed data. This data shows that SNR increases with increasing wavelength shift, to a peak at shift 5 (equivalent to 2.4 nm), before beginning to decrease. Within the SNR calculations, wavelength shift 5 provides the highest value for both sample types, for both types of SERDS spectra. It is also clear from this data that the use of the reconstructions provides higher SNR values than those obtained using the difference spectra.
This increase in SNR with increasing wavelength difference (until the peak at wavelength shift 5) occurs as a result of the greater separation between the peaks; less overlap between the peaks means that more of the intensity of the original spectrum at 830.0 nm is retained within the difference spectra. This also allows for a more efficient combination between the positive and negative sections of the split peaks, and thus a higher SNR. Similarly, the increase in signal within the reconstructed spectra results from the addition of the split peaks or first derivative line shapes to form one single reconstructed peak and is to be expected. However, for wavelength shifts 6 and 7, the reduction in Raman intensities associated with less efficient filtration when increasing the excitation wavelength outweighs the positive impact of increasing the separation of the peaks.
This result corroborates literature reports of an increase in the intensity of difference spectra peaks with increasing change in the two excitation wavelengths; the previously discussed 2017 study comparing wavelength shifts of 0.5, 1.0, 1.5 and 2.0 nm with a 784 nm excitation found that 2 nm shift gave the highest intensity values within the SERDS spectra.19 Furthermore, simulations within this work combined the resulting difference spectra from shifts ranging from 0 to 12.6 nm with auto-correlation calculations of pure Raman spectra and determined that for lipid and protein samples the optimal shifts were 7 and 10 nm, respectively. These shifts were identified as optimal because they retain a maximal amount of chemical information from the original signals, although the authors noted that owing to experimental constraints with optical filters, only shifts of up to 2 nm could be tested within their experimental setup. A 2015 study comparing 0.1, 0.3, 0.4, 0.5, 0.6, 0.9 and 1.0 nm shifts with a 785 nm excitation found that the 1 nm shift led to the highest peak intensities, when measuring human oral tissue.49 Moreover, a 2020 study by Korinth et al. noted that an excitation wavelength gap of 2 nm produced less noisy difference spectra than a 1 nm gap.50 The results presented here demonstrate a good agreement with these trends.
The next step in analysis was to calculate the signal (∼1453 cm−1 peak) to background ratio for the SERDS difference and reconstructed spectra as a function of the wavelength shift number. Whilst the predominant aim of this75 series of experiments is to determine the optimum excitation wavelength for the removal of fluorescence and etaloning signals, it can be seen in (Fig. 4a) that silica signals are present within the spectra (below ∼870 nm). It is therefore valuable to consider how the change in wavelength shifts affect the silica background removal within highly fluorescent samples. The ratio between the intensity of signals originating from the sample itself and the silica background region within the spectra serves as a metric for this analysis. The difference spectra results are plotted in Fig. 5b(i) and the reconstructions within Fig. 5b(ii).
It is clear from this data that there is a difference in trend by sample type, for the difference spectra. The bovine cheek values were higher than the porcine liver, although both samples have a peak at wavelength shift 4 (2.0 nm), with values of 5.63 and 2.97, respectively. The difference in trend is that for the bovine cheek samples, SBR values begin fairly high, peak at wavelength shift 4 and then begin decreasing, whereas for porcine liver, values begin low, increase to their peak and then remain fairly high until a large decrease at wavelength shift 7 (3.9 nm). For the cheek samples, shifts 2–4 are the ideal candidates, whereas for the liver samples, shifts 4–6 offer the best silica removal performance. For both types of samples, the sample peak intensity increases until wavelength shift 5 as previously described within the SNR analysis. However, within the cheek samples there is greater variation in the silica background region as a function of wavelength shift, and the success of the removal of the broad background region begins to decrease at wavelength shift 4. This results in a decrease to the SBR value at wavelength shift 5 and above. Nevertheless, the average SBR values for the 40 s, 830.0 nm excitation, standard Raman spectra of the bovine cheek and porcine liver samples were 0.32 ± 0.012 and 0.16 ± 0.009, respectively. Therefore, it is clear that the use of any of the SERDS difference spectra results in a significant reduction to the relative silica contributions (the minimum value obtained across all parameters is 0.97 ± 0.17, for porcine liver, wavelength shift 7).
For the reconstructions, wavelength shifts 5 and 6 optimise the removal of the broad silica signals with respect to the signal intensity of the peaks originating from within the sample. The SBR values obtained using the reconstructed spectra are higher than those obtained with the difference spectra, as a result of the increase in signal to the sample peaks. This effect appears to outweigh the changes to the silica region within the SERDS reconstructions, as both sample types now demonstrate optimal performance at wavelength shifts 5 and 6. Once again, the values are significantly higher than those obtained by the standard Raman spectra, indicating the strength of the technique for removing silica contributions.
As a final point of comparison the standard deviation of the 1782–1817 cm−1 region was used as a measure of levels of etaloning signals. The SBR values obtained by the standard Raman spectra of the bovine cheek and porcine liver samples were 173 ± 8 and 262 ± 13, respectively.
The results for the two types of SERDS spectra are shown in Fig. 5(c); subplot (i) demonstrates the difference spectra values, and subplot (ii) the reconstruction values. It is clear that the etaloning removal performance is fairly consistent across each wavelength shift, until shifts 6 and 7, with shift 7 demonstrating the overall worst performance. This is likely as a result of the reduction to overall intensity associated with the final two excitation wavelengths creating slight changes to the etaloning pattern and therefore resulting in a less faithful removal of the pattern, as well as relatively lower signals caused by laser wavelength reaching the bandwidth of the clean up filter.
The etaloning values obtained using the difference spectra are consistently higher than those obtained using the reconstructions, however, it is also clear from these results that the reconstructed spectra have higher SNR and lower noise levels than the SERDS difference spectra. Given that this method of evaluating levels of etaloning simply uses the standard deviation of the relevant region, it is possible that the etaloning values appear higher in the difference spectra due to the increased noise as compared to the simulations. Nevertheless, it is clear from this data that both types of SERDS spectra using wavelength shifts 1–5 significantly reduce the presence of etaloning as compared to standard Raman spectra.
In conclusion, for this set up measuring biological tissue samples, wavelength shift 5 (830.0 nm and 832.4 nm) is the best option for increasing overall data quality whilst also ensuring the most successful removal of etaloning contributions within the spectra. Given that shifts 1–5 all produce comparable etaloning removal, and that shift 6 shows lower etaloning removal efficiency, whilst shift 5 consistently performs well in other indicators of spectral quality, shift 5 is the best option. Although wavelength shift 5 does not produce optimal silica background removal in the bovine cheek difference spectra, the performance is still significantly higher than standard Raman spectroscopy, and in all other comparisons shift 5 gives strong (if not the best) results. Moreover, the primary motivation for this work is reducing the etaloning contributions, and as such, wavelength shift 5 represents the optimal choice.
This result demonstrates good agreement with literature reports that when measuring biological samples, it is beneficial to use large wavelength shifts. The benefit of increasing the wavelength is outweighed by the mismatch between the excitation wavelength and the filtration within the setup for shifts 6 and 7, resulting in shift 5 representing the optimal choice within this work.
In terms of the performance of the difference and reconstructed spectra, it is clear that the use of the reconstructed spectra increases the overall signal levels originating from the sample and therefore the signal to noise ratio, as well as reduces the amount of relative silica contributions within the spectra. It is important to consider that reconstruction of the SERDS difference spectra has the potential to introduce distortions into the dataset,51 however these results indicate even the SERDS difference spectra outperform the standard Raman data. This analysis demonstrated that the SERDS technique has strong potential to make even samples with extremely high levels of fluorescence measurable. It should be noted that the acquisition times within these experiments is higher than clinically applicable (20 s, 40 s), however, the needle probe #1 used to collect the data was a simple 1-in-1-out design. The use of a needle probe with a higher collection efficiency would enable a reduction to the acquisition time without sacrificing the data quality.
Lymph node measurements
The long-term aim of this work is to identify the optimal parameters for utilising the SERDS technique, in combination with Raman needle probes, for the assessment of highly fluorescent tissue in vivo. Therefore, it is valuable to test the method with human tissue samples to evaluate its potential. Wavelength shift 5 was used to measure 1 cancerous and 1 reactive (a benign inflammatory condition often mistaken for cancer76) lymph node, before standard Raman measurements were taken as a point of comparison. To the best of the authors’ knowledge, these represent the first measurements of human lymph node samples using the SERDS technique. Within these measurements, a different Raman needle probe design (‘Raman needle probe #2′, described within ‘Experimental methods – Raman probe designs’) was combined with the tuneable Ti:Sapphire laser setup, which allowed for a reduction to the acquisition times whilst maintaining good spectral quality.Fig. 6a(i) demonstrates the average raw spectra for both the reactive and cancerous lymph node, taken with 830.0 nm and 832.4 excitation. It is clear from the raw spectra that high levels of fluorescence and etaloning are present within both of the average spectra. Within the average cancerous spectrum sub Fig. a(i)), the etaloning can most clearly be seen within the ∼950–960 nm region, although it also appears from ∼965–980 nm. With 830 nm excitation, this corresponds to the 1521–1631 cm−1 and 1685–1844 cm−1 spectral regions. As well as these etaloning signals, both silica contributions and signals originating from within the sample can be seen within the average spectra. This is also true of the average reactive lymph node spectra, with the intensity of the peaks originating from the sample being higher, and the etaloning appearing to predominantly affect approximately the same spectral regions as in the cancerous spectra.
Fig. 6(b) demonstrates the average standard Raman (sub Fig. (i)), SERDS difference (sub Fig. (ii)) and SERDS reconstructed (sub Fig. (ii)) data for the reactive lymph node sample. It is clear from this data that the same chemical peaks are present within the each of the different types of spectra, including the peaks at 1083 cm−1, 1305 cm−1, 1442 cm−1 and 1660 cm−1. It is also clear that the use of the SERDS technique results in spectra that show no visible etaloning signals, and reduces the broad silica background contributions shown within the raw spectra whilst retaining chemical information. The standard Raman spectra show clear etaloning signals within the ∼1500–1640 cm−1 and 1790–1912 cm−1 spectral regions. These can impact negatively on spectral classification algorithms and should be minimised when possible.
This confirms the potential for SERDS to improve the data quality when measuring lymph node tissue. The prominent peaks within the average cancerous lymph node spectra (Fig. 6a(i)) of 1007 cm−1, 1305 cm−1, 1442 cm−1 and 1661 cm−1 are also conserved within both the SERDS difference and reconstructed spectra (not shown here but seen in ESI Fig. 3†), indicating that the SERDS technique maintains chemical peaks whilst also reducing levels of etaloning and silica signals within the spectra.
Whilst only one lymph node of each pathology was measured (and therefore limited conclusions regarding pathology dependent signals can be drawn); within Raman spectroscopy the spectral regions of 470–1200 cm−1, 1500–1700 cm−1, and the peaks of 980 cm−1, 1080 cm−1 and 1240 cm−1 are commonly associated with carbohydrates, proteins and the phosphate groups of DNA, respectively.1 More specifically, a 2013 study of lymph nodes within the head and neck identified a prominent peak at 1001 cm−1 within their data, attributed to phenylalanine within collagen.77 A 2019 study using Raman spectroscopy to investigate the differences between reactive and primary cancer lymph nodes in childhood Non-Hodgkin Lymphoma cases76 also identified strong peaks at 1006 cm−1, 1301 cm−1, 1443 cm−1 and 1667 cm−1, attributed to carotenoids, CH, CH2 and CH3 normal modes of lipid, protein and collagen, normal modes of lipids, fatty acids and proteins, and lipids and proteins, respectively. Both of these studies also reported the presence of the same peak locations across different pathology types for the lymph node tissue, with subtle variations in intensity by classification. A further 2020 study analysing both reactive and Non-Hodgkin Lymphoma samples also identified peaks at 1448 cm−1 and 1659 cm−1 as the most prominent within the spectra.78
It is therefore clear that the peaks shown within the spectra in Fig. 6 demonstrate good agreement with literature reports of the Raman peaks of lymph node tissue. It is noted within the 2013 study that the typical trend for cancerous samples is an increase in DNA contributions, owing to the dysregulated growth of cells, however, the lymphocytes contained within healthy lymph nodes contain strong nucleic acid signals and as such when metastatic invasion occurs the corresponding increase in epithelial cells causes an overall decrease to the relative nuclear to cytoplasmic ratio.77 Interestingly, the peak at 1083 cm−1 which is present within the reactive (and absent in the cancerous) lymph node spectra, has been assigned in previous studies to a C–C out of plane stretch73,79 of DNA.1 With only one sample of each pathology measured it is not possible to infer trends in chemical signals, however the apparent correlation between the average spectra and literature reports further demonstrates the potential of this technique.
In order to quantify the improvement in data quality generated through the use of the SERDS spectra, SNR, SBR and standard deviation of the higher wavenumber etaloning region (1717.3 cm−1–1753.5 cm−1) calculations were completed for each of the different types of spectra for both sample types. The peak at 1444 cm−1 was utilised for the ‘signal’ values in these calculations. The results of this analysis can be seen in Table 2.
| Sample | Spectra | SBR | SNR | Etaloning |
|---|---|---|---|---|
| Cancer | Standard | 0.239 ± 0.033 | 38.9 ± 13 | 6.04 ± 1.1 |
| Cancer | Difference | 2.89 ± 0.93 | 18.6 ± 10.6 | 5.70 ± 0.91 |
| Cancer | Reconstruct | 24 ± 2.6 | 164.2 ± 46.6 | 4.95 ± 1.23 |
| Reactive | Standard | 1.79 ± 1.41 | 280.1 ± 230 | 19.0 ± 9.0 |
| Reactive | Difference | 17 ± 11 | 155.4 ± 134.7 | 8.05 ± 2.73 |
| Reactive | Reconstruct | 28 ± 19 | 257.4 ± 195.6 | 6.79 ± 2.58 |
When considering these values, it is clear to see that the use of the difference spectra improves the relative silica contributions to the spectra, and that the use of the reconstructions further improves this trend. The improvement conferred by the use of the reconstructions is significantly higher within the cancerous samples. This is likely due to the fact that the peak intensities within the standard spectra of the reactive samples are already relatively high with comparison to the silica signals. Therefore, the improvement is not as significant as within the cancerous sample measurements, where the sample contributions are smaller with respect to the silica samples. It should be noted here that the SBR values for the standard Raman spectra were calculated using processed (i.e. baselined and SNV normalised) spectra, rather than representing the SBR for raw ‘standard’ Raman spectra.
The average SNR values for the cancerous sample data demonstrates that the use of the difference spectra reduces the achievable SNR, whereas the reconstructed spectra achieve a much higher SNR than either the standard or difference spectra. As previously mentioned, the SNR of the reconstructed spectra is expected to be higher than that of the difference spectra owing to the removal of fixed pattern noise and combination of the two halves of the split peaks.62 It has also been identified within the literature that the use of the difference spectra results in a SNR reduction when compared with standard Raman spectra of the same overall acquisition time as the difference spectra. This is as a result of both the subtraction of the peak intensities from one another, and the addition of the noise of each of the individual spectra.52 The values for the reactive spectra once again demonstrate that the use of the difference spectra reduces the achievable SNR, however, the use of the SERDS reconstruction does increase the performance with respect to the difference spectra, but not with respect to the standard Raman spectra. Once again, this is likely due to the fact that the standard Raman spectra of the reactive sample demonstrate much higher intensity levels than the cancerous sample, and as such the SERDS technique does not offer an improvement to overall signal to noise ratio. This does however demonstrate the potential of SERDS to offer a significant improvement to data quality within samples with lower Raman signals.
It can also be seen within these values that the standard deviation of the average SNR values is much higher within the reactive sample spectra than for the cancerous. This is because there is a large amount of variation within the spectra obtained from this sample. To illustrate, Fig. 7 demonstrates the average baselined and normalised standard Raman spectra from the two main ‘types’ of spectral signal generated from the reactive lymph node sample. It is clear from these spectra that some measurements from the sample demonstrate spectral signatures similar to those obtained with adipose tissue, and that others show higher levels of fluorescence, inducing etaloning and some different Raman peak positions and intensities. It is well established within literature that lymph nodes are often located within, or attached to, adipose tissue, and that lymph fluid contains high concentrations of emulsified fats.31 This large variation in spectral signatures from within the sample is not present within the cancerous spectra and therefore accounts for this large increase in standard deviation.
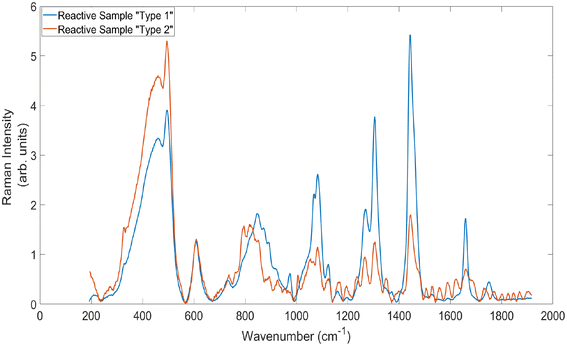 | ||
| Fig. 7 Average standard Raman spectra of the two main types of signals generated by measuring the reactive lymph node sample. Acquisition time = 5 × 20 s. | ||
The etaloning values obtained confirm the value of the SERDS technique for reducing etaloning. Both within the reactive and cancerous samples the use of the difference and reconstructed spectra results in a corresponding reduction to the standard deviation of the etaloning region. Once again, the difference between the SERDS and standard spectra are more pronounced within the reactive samples, which is likely due to the fact that the reactive samples had higher levels of etaloning to be removed.
The acquisition times utilised within these measurements (20 s total) are higher than would be required for use within in vivo analysis. However, the needle probe design within this setup (‘needle probe #2′) facilitated the use of lower acquisition times than the 1-in-1-out probe (‘needle probe #1′) used within the measurements of animal tissue. There is also significant further scope to increase the collection efficiency of the design. This would be expected to result in a reduction to the necessary acquisition times when measuring these highly fluorescent samples.
Overall, this data demonstrates that the SERDS technique, with an excitation wavelength shift of 2.4 nm, has great potential for the removal of both etaloning and broad silica background contributions to the spectra. Moreover, the use of the technique retains the chemical signals originating from within the sample. Whilst the use of the difference spectra reduces the achievable SNR, the reduction in silica and particularly etaloning represents an overall improvement to data quality. Moreover, the reconstructed SERDS spectra can actually result in an improvement to the achievable SNR (as compared to both conventional Raman and SERDS difference spectra) when measuring samples with low levels of scattering. These results highlight the powerful potential of the technique.
As a final step in the analysis, principal components analysis (PCA) was performed on the spectra from both the cancerous and reactive lymph node, using both the standard Raman spectra, and both types of the SERDS spectra. Given that only one sample of each pathological type was measured within this work, it is not possible to create a diagnostically accurate or statistically relevant model using this data. However, given the ability of PCA to identify statistical variance within a dataset, the comparison between the standard Raman and SERDS reconstructed spectra can elucidate whether etaloning is a significant source of this variance for each data set.
The results of this analysis, along with a more in depth discussion, can be seen in ESI Fig. 4–6.† The data clearly demonstrates that the use of both of the SERDS spectra results in a stronger statistical separation between the two pathology types than conventional spectra taken using the Raman needle probe #2. Most importantly, the use of the SERDS spectra removes the presence of etaloning from within the most significant principal components, indicating that the highest levels of variation originate instead from chemical changes from within the sample. The loadings within these basic PCA models also illustrate that the use of the SERDS technique results in a reduction to the luminescent silica contributions.
Conclusions
Within these experiments, 7 different SERDS wavelength shifts (between 0.4 and 3.9 nm) were evaluated for their impact upon data quality; in terms of fluorescence suppression, signal to noise ratio and removal of silica and etaloning spectral contributions. Comparison between SERDS spectra and spectra taken using a Raman microscope confirmed that the SERDS spectra were able to retrieve the same chemical signals as are present within ‘typical’ Raman spectra of tissue. Testing using highly fluorescent porcine liver and bovine cheek samples indicated that a combination of 830.0 and 832.4 nm excitation provided the highest quality data, and significantly reduced the levels of etaloning obtained within the spectra. This excitation wavelength shift was then utilised to measure one cancerous and one reactive ex vivo lymph node sample, using our more advanced #2 needle probe being translated for in vivo applications. These preliminary proof of concept measurements demonstrated a significant improvement in performance (a reduction in fluorescence, silica, and etaloning levels), and a subsequent improvement in PCA discrimination when using the SERDS spectra.Author contributions
Dr Hannah Sheridan was responsible for data collection and curation, formal analysis, investigation and writing of the original draft of the manuscript. Drs Alexander Dudgeon and John Day were responsible for review and editing the manuscript. Dr Catherine Kendall contributed resources, as well as reviewing and editing of the manuscript. Mr Charles Hall contributed to the sample collection and data curation and resources of the project. Prof. Nick stone was responsible for the conceptualisation, funding acquisition, and supervision, as well as writing, reviewing and editing of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors wish to thank all the staff in the department of Otolaryngology and Head & Neck Surgery, Gloucestershire Hospital. This project is funded by the NIHR i4i (II-LB-1117-20002) and a UKRI EPSRC doctoral training partnership funded the PhD studentship of H. Sheridan, who is the first author.References
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis and J. Dorney, et al., Using Raman spectroscopy to characterize biological materials, Nat. Protoc., 2016, 11(4), 664–687, DOI:10.1038/nprot.2016.036.
- M. G. Ramírez-Elías and F. J. González, Raman Spectroscopy for In Vivo Medical Diagnosis, in Raman Spectroscopy, InTech, 2018. Available from: https://www.intechopen.com/books/raman-spectroscopy/raman-spectroscopy-for-in-vivo-medical-diagnosis Search PubMed.
- K. Kong, C. Kendall, N. Stone and I. Notingher, Raman spectroscopy for medical diagnostics — From in-vitro biofluid assays to in-vivo cancer detection, Adv. Drug Delivery Rev., 2015, 89, 121–134 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0169409X15000447.
- G. W. Auner, S. K. Koya, C. Huang, B. Broadbent, M. Trexler and Z. Auner, et al., Applications of Raman spectroscopy in cancer diagnosis, Cancer Metastasis Rev., 2018, 37(4), 691 CrossRef CAS PubMed . Available from: https://pmc/articles/PMC6514064/.
- S. P. Mulvaney and C. D. Keating, Raman Spectroscopy, Anal. Chem., 2000, 72(12), 145–157 CrossRef CAS PubMed.
- A. Kudelski, Analytical applications of Raman spectroscopy, Talanta, 2008, 76(1), 1–8 CrossRef CAS PubMed.
- M. Jermyn, K. Mok, J. Mercier, J. Desroches, J. Pichette and K. Saint-Arnaud, et al., Intraoperative brain cancer detection with Raman spectroscopy in humans, Sci. Transl. Med., 2015, 7(274), 274ra19 CAS . Available from: https://www.ncbi.nlm.nih.gov/pubmed/25673764.
- H. F. Nargis, H. Nawaz, H. N. Bhatti, K. Jilani and M. Saleem, Comparison of surface enhanced Raman spectroscopy and Raman spectroscopy for the detection of breast cancer based on serum samples, Spectrochim. Acta, Part A, 2021, 246, 119034 CrossRef CAS PubMed.
- H. Lui, J. Zhao, D. McLean and H. Zeng, Real-time Raman spectroscopy for in vivo skin cancer diagnosis, Cancer Res., 2012, 72(10), 2491–2500 CrossRef CAS PubMed . Available from: https://www.ncbi.nlm.nih.gov/pubmed/22434431.
- H. C. McGregor, M. A. Short, A. McWilliams, T. Shaipanich, D. N. Ionescu and J. Zhao, et al., Real-time endoscopic Raman spectroscopy for in vivo early lung cancer detection, J. Biophotonics, 2017, 10(1), 98–110, DOI:10.1002/jbio.201500204.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Raman spectroscopy for identification of epithelial cancers, Faraday Discuss., 2004, 126, 141–57 RSC . Available from: https://rsc.66557.net/en/content/articlehtml/2004/fd/b304992b.
- A. Lagree, A. Shiner, M. A. Alera, L. Fleshner, E. Law and B. Law, et al., Assessment of digital pathology imaging biomarkers associated with breast cancer histologic grade, Curr. Oncol., 2021, 28(6), 4298–4316 CrossRef PubMed . Available from: https://pmc/articles/PMC8628688/.
- P. M. Speight, T. J. Abram, P. N. Floriano, R. James, J. Vick and M. H. Thornhill, et al., Interobserver agreement in dysplasia grading: toward an enhanced gold standard for clinical pathology trials, Oral Surg. Oral Med. Oral Pathol. Oral Radiol., 2015, 120(4), 474–482 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26216170/.
- K. H. Yu, C. Zhang, G. J. Berry, R. B. Altman, C. Ré and D. L. Rubin, et al., Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features, Nat. Commun., 2016, 7(1), 1–10 Search PubMed . Available from: https://www.nature.com/articles/ncomms12474.
- C. E. L. Klaver, N. Bulkmans, P. Drillenburg, H. I. Grabsch, N. C. T. van Grieken and A. Karrenbeld, et al., Interobserver, intraobserver, and interlaboratory variability in reporting pT4a colon cancer, Virchows Arch., 2020, 476(2), 219–230, DOI:10.1007/s00428-019-02663-0.
- J. M. Bueno-de-Mesquita, D. S. A. Nuyten, J. Wesseling, H. van Tinteren, S. C. Linn and M. J. van de Vijver, The impact of inter-observer variation in pathological assessment of node-negative breast cancer on clinical risk assessment and patient selection for adjuvant systemic treatment, Ann. Oncol., 2010, 21(1), 40–47 CrossRef CAS PubMed . Available from: https://linkinghub.elsevier.com/retrieve/pii/S0923753419382419.
- K. Ranganathan, L. Kavitha, P. Sharada, R. M. Bavle, R. S. Rao and S. M. Pattanshetty, et al., Intra-observer and inter-observer variability in two grading systems for oral epithelial dysplasia: A multi-centre study in India, J. Oral. Pathol. Med., 2020, 49(9), 948–955, DOI:10.1111/jop.13056.
- C. Kendall, N. Stone, N. Shepherd, K. Geboes, B. Warren and R. Bennett, et al., Raman spectroscopy, a potential tool for the objective identification and classification of neoplasia in Barrett's oesophagus, J. Pathol., 2003, 200(5), 602–609 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/12898596/.
- E. Cordero, I. Latka, C. Matthäus, I. W. Schie and J. Popp, In-vivo Raman spectroscopy: from basics to applications, J. Biomed. Opt., 2018, 23(07), 1, DOI:10.1117/1.JBO.23.7.071210.full.
- I. Pence and A. Mahadevan-Jansen, Clinical instrumentation and applications of Raman spectroscopy, Chem. Soc. Rev., 2016, 45(7), 1958–79 RSC . Available from: https://xlink.rsc.org/?=C5CS00581G.
- J. Desroches, M. Jermyn, M. Pinto, F. Picot, M. A. Tremblay and S. Obaid, et al., A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy, Sci. Rep., 2018, 8(1), 1792 CrossRef PubMed . Available from: https://www.ncbi.nlm.nih.gov/pubmed/29379121.
- R. O. P. Draga, M. C. M. Grimbergen, P. L. M. Vijverberg, C. F. P. van Swol, T. G. N. Jonges and J. A. Kummer, et al., In Vivo Bladder Cancer Diagnosis by High-Volume Raman Spectroscopy, Anal. Chem., 2010, 82(14), 5993–5999, DOI:10.1021/ac100448p.
- P. Crow, A. Molckovsky, N. Stone, J. Uff, B. Wilson and L. M. WongKeeSong, Assessment of fiberoptic near-infrared raman spectroscopy for diagnosis of bladder and prostate cancer, Urology, 2005, 65(6), 1126–1130 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0090429504015328.
- W. Wang, J. Zhao, M. Short and H. Zeng, Real-time in vivo cancer diagnosis using raman spectroscopy, J. Biophotonics, 2015, 8(7), 527–545, DOI:10.1002/jbio.201400026.
- L. M. Almond, J. Hutchings, G. Lloyd, H. Barr, N. Shepherd and J. Day, et al., Endoscopic Raman spectroscopy enables objective diagnosis of dysplasia in Barrett's esophagus, Gastrointest. Endosc., 2014, 79(1), 37–45 CrossRef PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0016510713019871.
- S. Coda, P. Siersema, G. Stamp and A. Thillainayagam, Biophotonic endoscopy: a review of clinical research techniques for optical imaging and sensing of early gastrointestinal cancer, Endosc. Int. Open, 2015, 03(05), E380–E392, DOI:10.1055/s-0034-1392513.
- A. Malpica, A. Mahadevan-Jansen, D. L. Heintzelman, M. Follen, R. Richards-Kortum and U. Utzinger, Near-Infrared Raman Spectroscopy for in Vivo Detection of Cervical Precancers, Appl. Spectrosc., 2001, 55(8), 955–959 CrossRef . Available from: https://www.osapublishing.org/as/abstract.cfm?uri=as-55-8-955.
- A. Mahadevan-Jansen, M. F. Mitchell, N. Ramanujam, U. Utzinger and R. Richards-Kortum, Development of a Fiber Optic Probe to Measure NIR Raman Spectra of Cervical Tissue In Vivo, Photochem. Photobiol., 1998, 68(3), 427–431, DOI:10.1111/j.1751-1097.1998.tb09703.x.
- A. Saha, I. Barman, N. C. Dingari, L. H. Galindo, A. Sattar and W. Liu, et al., Precision of Raman Spectroscopy Measurements in Detection of Microcalcifications in Breast Needle Biopsies, Anal. Chem., 2012, 84(15), 6715–6722, DOI:10.1021/ac3011439.
- R. Sathyavathi, A. Saha, J. S. Soares, N. Spegazzini, S. McGee and R. Rao Dasari, et al., Raman spectroscopic sensing of carbonate intercalation in breast microcalcifications at stereotactic biopsy, Sci. Rep., 2015, 5(1), 9907 CrossRef CAS PubMed . Available from: https://www.nature.com/articles/srep09907.
- L. M. Fullwood, I. E. Iping Petterson, A. P. Dudgeon, G. R. Lloyd, C. Kendall and C. Hall, et al., Evaluation of a multi-fibre needle Raman probe for tissue analysis, in Biomedical Vibrational Spectroscopy 2016: Advances in Research and Industry, ed. A. Mahadevan-Jansen and W. Petrich, International Society for Optics and Photonics, 2016, p. 97040G. DOI:10.1117/12.2230005.
- I. E. Iping Petterson, J. C. C. Day, L. M. Fullwood, B. Gardner and N. Stone, Characterisation of a fibre optic Raman probe within a hypodermic needle, Anal. Bioanal. Chem., 2015, 407(27), 8311–8320, DOI:10.1007/s00216-015-9021-7.
- M. Jermyn, J. Desroches, K. Aubertin, K. St-Arnaud, W. J. Madore and E. De Montigny, et al., A review of Raman spectroscopy advances with an emphasis on clinical translation challenges in oncology, Phys. Med. Biol., 2016, 61(23), R370–R400 CrossRef CAS PubMed . Available from: https://stacks.iop.org/0031-9155/61/i=23/a=R370?key=crossref.c406f22d1b9e70b37ade780e496ebf66.
- O. Stevens, I. E. Iping Petterson, J. C. C. Day and N. Stone, Developing fibre optic Raman probes for applications in clinical spectroscopy, Chem. Soc. Rev., 2016, 45(7), 1919–1934 RSC . Available from: https://xlink.rsc.org/?DOI:=C5CS00850F.
- C. Kallaway, L. M. Almond, H. Barr, J. Wood, J. Hutchings and C. Kendall, et al., Advances in the clinical application of Raman spectroscopy for cancer diagnostics, Photodiagn. Photodyn. Ther., 2013, 10(3), 207–219 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S1572100013000124.
- D. Wei, S. Chen and Q. Liu, Review of Fluorescence Suppression Techniques in Raman Spectroscopy, Appl. Spectrosc. Rev., 2015, 50(5), 387–406, DOI:10.1080/05704928.2014.999936.
- R. Li, D. Verreault, A. Payne, C. L. Hitchcock, S. P. Povoski and E. W. Martin, et al., Effects of laser excitation wavelength and optical mode on Raman spectra of human fresh colon, pancreas, and prostate tissues, J. Raman Spectrosc., 2014, 45(9), 773–780, DOI:10.1002/jrs.4540.
- F. X. Liu, Y. Xiao and Y. S. Li, Near-infrared raman spectroscopy of human lung tissues: Possibility of molecular-level cancer diagnosis, J. Raman Spectrosc., 2001, 32(2), 139–141 CrossRef.
- J. C. C. Day and N. Stone, A Subcutaneous Raman Needle Probe, Appl. Spectrosc., 2013, 67(3), 349–354, DOI:10.1366/12-06651.
- R. E. Kast, G. K. Serhatkulu, A. Cao, A. K. Pandya, H. Dai and J. S. Thakur, et al., Raman spectroscopy can differentiate malignant tumors from normal breast tissue and detect early neoplastic changes in a mouse model, Biopolymers, 2008, 89(3), 235–241, DOI:10.1002/bip.20899.
- R. E. Kast, S. C. Tucker, K. Killian, M. Trexler, K. V. Honn and G. W. Auner, Emerging technology: Applications of Raman spectroscopy for prostate cancer, Cancer Metastasis Rev., 2014, 33(2–3), 673–693, DOI:10.1007/s10555-013-9489-6.
- HORIBA. Horiba Scientific. 2022. Selecting a CCD Camera for Spectroscopic Applications. Available from: https://www.horiba.com/int/ccd-camera-spectroscopic-applications/.
- R. Heintz, Back illuminated vs. front illuminated CCD-based imaging sensors and how it impacts Raman spectra, Waltham, Massachusetts, 2019 Search PubMed.
- Teledyne Imaging, How a Charge Coupled Device (CCD) Image Sensor Works, Thousand Oaks, California, 2020 Search PubMed.
- C. Massie, K. Chen and A. J. Berger, Calibration Technique for Suppressing Residual Etalon Artifacts in Slit-Averaged Raman Spectroscopy, Appl. Spectrosc., 2022, 76(2), 255–61 CrossRef CAS PubMed.
- A. C. De Luca, K. Dholakia and M. Mazilu, Modulated Raman Spectroscopy for Enhanced Cancer Diagnosis at the Cellular Level, Sensors, 2015, 15(6), 13680–13704 CrossRef CAS PubMed . Available from: https://www.mdpi.com/1424-8220/15/6/13680/htm.
- K. Sowoidnich, M. Towrie and P. Matousek, Shifted Excitation Raman Difference Spectroscopy Combined with Wide Area Illumination and Sample Rotation for Wood Species Classification, Appl. Spectrosc., 2023, 77(6), 666–681 CrossRef CAS PubMed . Available from: https://opg.optica.org/abstract.cfm?uri=as-77-6-666.
- B. C. Wilson, M. Jermyn and F. Leblond, Challenges and opportunities in clinical translation of biomedical optical spectroscopy and imaging, J. Biomed. Opt., 2018, 23(3), 030901, DOI:10.1117/1.JBO.23.3.030901.full.
- M. T. Gebrekidan, C. Knipfer, F. Stelzle, J. Popp, S. Will and A. Braeuer, A shifted-excitation Raman difference spectroscopy (SERDS) evaluation strategy for the efficient isolation of Raman spectra from extreme fluorescence interference, J. Raman Spectrosc., 2016, 47(2), 198–209 CrossRef CAS.
- F. Korinth, A. S. Mondol, C. Stiebing, I. W. Schie, C. Krafft and J. Popp, New methodology to process shifted excitation Raman difference spectroscopy data: a case study of pollen classification, Sci. Rep., 2020, 10(1), 1–12 CrossRef PubMed . Available from: https://www.nature.com/articles/s41598-020-67897-4.
- F. Korinth, T. A. Shaik, J. Popp and C. Krafft, Assessment of shifted excitation Raman difference spectroscopy in highly fluorescent biological samples, Analyst, 2021, 146(22), 6760–6767 RSC . Available from: https://rsc.66557.net/en/content/articlehtml/2021/an/d1an01376a.
- E. Cordero, F. Korinth, C. Stiebing, C. Krafft, I. W. Schie and J. Popp, Evaluation of Shifted Excitation Raman Difference Spectroscopy and Comparison to Computational Background Correction Methods Applied to Biochemical Raman Spectra, Sensors, 2017, 17(8), 1724 CrossRef PubMed . Available from: https://www.mdpi.com/1424-8220/17/8/1724/htm.
- S. Dochow, N. Bergner, C. Matthäus, B. B. Praveen, P. C. Ashok and M. Mazilu, et al., Etaloning, fluorescence and ambient light suppression by modulated wavelength Raman spectroscopy, Biomed. Spectrosc. Imaging, 2012, 1(4), 383–389, DOI:10.3233/BSI-120031.
- S. Mosca, K. Sowoidnich, M. Mehta, W. H. Skinner, B. Gardner and F. Palombo, et al., 10 kHz Shifted-Excitation Raman Difference Spectroscopy with Charge-Shifting Charge-Coupled Device Read-Out for Effective Mitigation of Dynamic Interfering Backgrounds, Appl. Spectrosc., 2023, 77, 569–582, DOI:10.1177/00037028231167441.
- A. P. Shreve, N. J. Cherepy and R. A. Mathies, Effective Rejection of Fluorescence Interference in Raman Spectroscopy Using a Shifted Excitation Difference Technique, Appl. Spectrosc., 1992, 46(4), 707–711, DOI:10.1366/0003702924125122.
- M. Maiwald, A. Müller, B. Sumpf and G. Tränkle, A portable shifted excitation Raman difference spectroscopy system: device and field demonstration, J. Raman Spectrosc., 2016, 47(10), 1180–1184, DOI:10.1002/jrs.4953.
- L. Y. Zhao, G. P. Wu and Y. Z. Wu, Evaluation of Shifted-Excitation Raman Difference Spectroscopy and Comparison to Ordinary Portable Raman Spectroscopy in Identification of Seized Drugs, Spectroscopy, 2023, 38(8), 12–21 CAS.
- K. Sowoidnich, S. Vogel, M. Maiwald and B. Sumpf, Determination of Soil Constituents Using Shifted Excitation Raman Difference Spectroscopy, Appl. Spectrosc., 2022, 76(6), 712–722 CrossRef CAS PubMed.
- M. Maiwald, K. Sowoidnich and B. M. Sumpf, Pilot investigations on solids, liquids and gases using a portable shifted excitation Raman difference spectroscopy sensor system, in Plasmonics in Biology and Medicine, ed. V. D. Tuan, A. H. Ho-Pui and R. Krishanu, SPIE, 2023, DOI:10.1117/12.2649696.full.
- S. Innocenti, D. Q. Balbas, L. Pezzati, R. Fontana and J. Striova, Portable Sequentially Shifted Excitation Raman Spectroscopy to Examine Historic Powders Enclosed in Glass Vials, Sensors, 2022, 22(9), 3560 CrossRef CAS PubMed . Available from: https://www.mdpi.com/1424-8220/22/9/3560/htm.
- M. Maiwald, K. Sowoidnich and B. Sumpf, Pilot investigations on solids, liquids and gases using a portable shifted excitation Raman difference spectroscopy sensor system, Plasmon. Biol. Med., 2023, 12396, 1239602, DOI:10.1117/12.2649696.full.
- M. A. D. S. Martins, E. A. P. dos Santos, A. Fontes, H. D. S. Martinho, A. A. Martin and D. G. Ribeiro, Shifted-excitation Raman difference spectroscopy for in vitro and in vivo biological samples analysis, Biomed. Opt. Express, 2010, 1(2), 617–626 CrossRef PubMed . Available from: https://opg.optica.org/viewmedia.cfm?uri=boe-1-2-617&seq=0&html=true.
- B. Lorenz, S. Guo, C. Raab, P. Leisching, T. Bocklitz and P. Rösch, et al., Comparison of conventional and shifted excitation Raman difference spectroscopy for bacterial identification, J. Raman Spectrosc., 2022, 53(7), 1285–1292, DOI:10.1002/jrs.6360.
- Z. Han, B. D. Strycker, B. Commer, K. Wang, B. D. Shaw and M. O. Scully, et al., Characterization and Identification of Fungal Conidia via Shifted Excitation Raman Difference Spectroscopy, Adv. Phys. Sci., 2022, 06, 2240005 CrossRef . Available from: https://www.worldscientific.com.
- A. Lux, M. Realini, A. Botteon, M. Maiwald, A. Muller and B. Sumpf, et al., Advanced portable micro-SORS prototype coupled with SERDS for heritage science, Analyst, 2024, 149, 2317–2327 RSC.
- M. Maiwald, A. Müller, B. Sumpf, G. Erbert and G. Tränkle, Capability of shifted excitation Raman difference spectroscopy under ambient daylight, Appl. Opt., 2015, 54(17), 5520 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26192855/.
- J. Lin, D. Lin, S. Qiu, Z. Huang, F. Liu and W. Huang, et al., Shifted-excitation Raman difference spectroscopy for improving in vivo detection of nasopharyngeal carcinoma, Talanta, 2023, 257, 124330 CrossRef CAS PubMed.
- F. S. A. de Siqueira e Oliveira, A. M. da Silva, M. T. T. Pacheco, H. E. Giana and L. Silveira, Biochemical characterization of pathogenic bacterial species using Raman spectroscopy and discrimination model based on selected spectral features, Lasers Med. Sci., 2021, 36(2), 289–302, DOI:10.1007/s10103-020-03028-9.
- M. H. Ahmed, J. A. Byrne and W. Ahmed, Characteristic of silicon doped diamond like carbon thin films on surface properties and human serum albumin adsorption, Diamond Relat. Mater., 2015, 55, 108–116 CrossRef CAS.
- C. Gullekson, L. Lucas, K. Hewitt and L. Kreplak, Surface-Sensitive Raman Spectroscopy of Collagen I Fibrils, Biophys. J., 2011, 100(7), 1837–1845 CrossRef CAS PubMed.
- M. Z. Vardaki and N. Kourkoumelis, Tissue Phantoms for Biomedical Applications in Raman Spectroscopy: A Review, Biomed. Eng. Comput. Biol., 2020, 11, 1–15, DOI:10.1177/1179597220948100.
- U. Böcker, R. Ofstad, Z. Wu, H. C. Bertram, G. D. Sockalingum and M. Manfait, et al., Revealing covariance structures in fourier transform infrared and Raman microspectroscopy spectra: A study on pork muscle fiber tissue subjected to different processing parameters, Appl. Spectrosc., 2007, 61(10), 1032–1039 CrossRef PubMed.
- M. Motoyama, I. Nakajima, H. Ohmori, G. Watanabe and K. Sasaki, A Raman Spectroscopic Method of Evaluating Fat Crystalline States and Its Application in Detecting Pork Fat, JARQ, 2018, 52(1), 17–22 CrossRef CAS . Available from: https://www.jircas.go.jp.
- M. Gniadecka, P. A. Philipsen, S. Wessel, R. Gniadecki, H. C. Wulf and S. Sigurdsson, et al., Melanoma Diagnosis by Raman Spectroscopy and Neural Networks: Structure Alterations in Proteins and Lipids in Intact Cancer Tissue, J. Invest. Dermatol., 2004, 122(2), 443–449 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0022202X15306746?via%3Dihub.
- F. Ensani, S. Mehravaran, G. Irvanlou, M. Aghaipoor, S. Vaeli and E. Hajati, et al., Fine-needle aspiration cytology and flow cytometric immunophenotyping in diagnosis and classification of non-Hodgkin lymphoma in comparison to histopathology, Diagn. Cytopathol., 2012, 40(4), 305–310, DOI:10.1002/dc.21561.
- J. V. Rau, F. Marini, M. Fosca, C. Cippitelli, M. Rocchia and A. Di Napoli, Raman spectroscopy discriminates malignant follicular lymphoma from benign follicular hyperplasia and from tumour metastasis, Talanta, 2019, 194, 763–770 CrossRef CAS PubMed.
- G. R. Lloyd, L. E. Orr, J. Christie-Brown, K. McCarthy, S. Rose and M. Thomas, et al., Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis, Analyst, 2013, 138(14), 3900 RSC . Available from: https://xlink.rsc.org/?DOI=c2an36579k.
- M. Agsalda-Garcia, T. Shieh, R. Souza, N. Kamada, N. Loi and R. Oda, et al., Raman-Enhanced Spectroscopy (RESpect) Probe for Childhood Non-Hodgkin Lymphoma, SciMed. J., 2020, 2(1), 1 CrossRef PubMed . Available from: https://pmc/articles/PMC8172049/.
- L. B. Lyndgaard, K. M. Sørensen, F. Van Den Berg and S. B. Engelsen, Depth profiling of porcine adipose tissue by Raman spectroscopy, J. Raman Spectrosc., 2012, 43(4), 482–489 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an01264j |
| This journal is © The Royal Society of Chemistry 2025 |