Open Access Article
Open Access ArticleTwo-dimensional MXene enabled carbon quantum dots@Ag with enhanced catalytic activity towards the reduction of p-nitrophenol†
Yingxin Chena,
Chunli Yanga,
Xiaotong Huang a,
Lu Lia,
Na Yua,
Huan Xiea,
Zebin Zhua,
Yong Yuan
a,
Lu Lia,
Na Yua,
Huan Xiea,
Zebin Zhua,
Yong Yuan b and
Lihua Zhou
b and
Lihua Zhou *a
*a
aSchool of Biomedical and Pharmaceutical Sciences, Guangdong University of Technology, Guangzhou 510006, P.R. China. E-mail: qhzhoulh@gdut.edu.cn
bSchool of Environmental Science and Engineering, Guangdong University of Technology, Guangzhou, 510006, P.R. China
First published on 10th February 2022
Abstract
A composite of cuttlefish ink-based carbon quantum dots@Ag/MXene (CQD@Ag/MXene) was firstly synthesized by solvothermal method as a catalyst for reduction of p-nitrophenol (PNP) to p-aminophenol (PAP). CQD@Ag/MXene was characterized by scanning electron microscopy (SEM), field emission transmission electron microscopy (TEM), X-ray diffraction (XRD), X-ray photoelectron spectroscopy (XPS) and Raman. The results show that loading on 2D material MXene can prevent the aggregation of CQD@Ag and expose more active sites, which contributes to a superior catalytic activity with a pseudo-first-order rate constant k (2.28 × 10−2 s−1) and mass-normalized rate constant km (5700 s−1 g−1), nearly 2 times higher than CQD@Ag without MXene (k = 1.09 × 10−2 s−1 and km = 2725 s−1 g−1). Besides, CQD@Ag/MXene showed excellent reusability which even retained about 65% activity in successive 10 cycles. The high adsorption rate to PNP and the promotion of forming H radicals may be the reason for the outstanding catalytic activity of CQD@Ag/MXene. CQD@Ag/MXene can be a potential candidate in the removal of environmental pollutants due to its facile synthesis and high catalytic efficiency.
Introduction
p-Nitrophenol (PNP) is widely used in the fields of pharmaceuticals, pesticides, explosives, dyes and leather processing.1,2 Extensive research has demonstrated that PNP is carcinogenic and genetically toxic to humans and wildlife due to the persistency in aquatic environments and soils.3,4 Different approaches including photocatalytic degradation,5 electrochemical oxidation,6 adsorption7 and catalytic hydrogenation8 have been developed for removing or reducing environmental toxicity of PNP from aqueous media. Notably, p-aminophenol (PAP), the reduced product of PNP, low cytotoxicity, is a crucial chemical raw material with high commercial value.9 A common method of reducing PNP to PAP is catalytic hydrogenation, in which the catalyst is an essential factor. Catalytic hydrogenation technique with precious metal (e.g., Au, Ag, Pt, Pd)-based nanoparticles (NPs) as catalysts is most effective in reduction of nitro compound in the presence of NaBH4 which acts as reducing agent.10,11 Among these precious metals, Ag has attracted particular attention due to its lowest cost.12 However, single Ag NPs as a catalyst failed to achieve the expected catalytic effect because of its easy oxidation, aggregation and poor stability.13In order to improve catalytic performance, the researchers incorporated Ag NPs into various substrates, such as polymers,14 carbons,15 porous organic frameworks (POFs),16 etc. to synthesize composites. Carbon materials including graphene, carbon nanotubes (CNTs) and carbon quantum dots (CQDs) attract the attention of researchers due to their high specific surface area, tunable porosity, easy functionalization, high chemical and thermal stability.17,18 Among them, CQDs as a new type of carbon based zero-dimensional functional material can reduce metal-salts to synthesize corresponding metal nanocomposites which exhibit the advantages of each single component and show off superior catalytic performance.19 Ghosh et al. reported a novel approach for synthesis of Au NPs by using CQDs as reducing as well as stabilizing agent, which can enhance catalytic activity towards reduction of PNP.20 Although nanocomposites have better catalytic properties than NPs, agglomeration occurs during the catalytic process due to their high surface energy, affecting catalytic activity and experimental reproducibility.21
The newly-fashioned 2D layered inorganic materials, such as silicene, black phosphorus (BP), transition metal oxides (TMOs), graphitic carbon nitride (g-C3N4), and MXenes etc., have attracted a lot of attention due to their unique properties and important applications.22,23 Among these 2D materials, MXene exhibits a unique 2D layered structure with a large specific surface area, good electrical conductivity, mechanical flexibility, chemical and thermal stability, environment-friendly characteristics, and other properties.24,25 MXene is a transition metal carbide or carbonitride produced in the HF solution by exfoliating the “A” layered component of the MAX matrix phase, which combines the advantages of ceramics and metals, and has a wide applications in water treatment,26 catalysis,27 adsorption,28 and new polymer reinforced composites.29,30 Zhang et al. had reported that MXene as a support of single Pt atom to enhance the catalytic activity for the hydrogen evolution reaction, and the catalytic activity exceeded the most advanced commercial HER catalyst Pt/C.31 Hence, MXene seems to be an ideal support to reduce the accumulation of nanocomposites during the catalysis process.
In this study, CQD@Ag/MXene as a new catalyst for the reduction reaction of PNP was synthesized, characterized, and evaluated. Scheme 1 shows the fabrication of CQD@Ag/MXene nanoparticles and the reduction mechanism of PNP in the presence of NaBH4. CQD@Ag was synthesized with the CQDs which was prepared from cuttlefish juice as a reducing agent, then was loaded on the 2D material MXene to obtain CQD@Ag/MXene. The morphology, elemental composition and group analysis of CQD@Ag and CQD@Ag/MXene were assayed. The catalytic activity and reusability of the CQD@Ag and CQD@Ag/MXene were studied using the reduction of an environmental pollutant PNP as a model reaction.
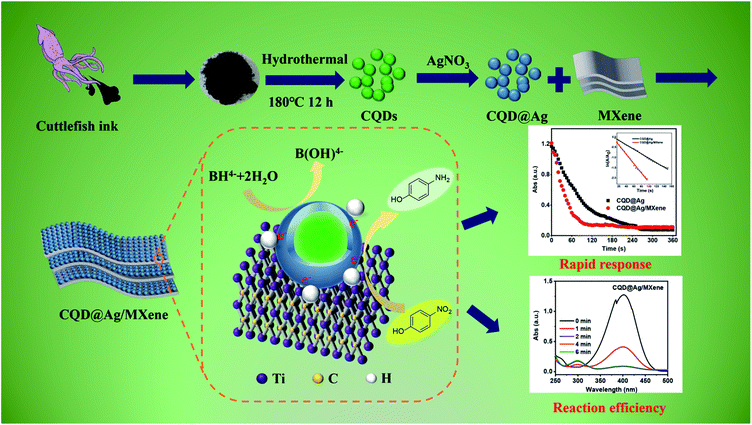 | ||
| Scheme 1 Schematic illustration of the fabrication of CQD@Ag/MXene and the reduction of PNP in the presence of NaBH4. | ||
Experimental
Materials
Ag NPs, AgNO3, PNP and 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) were all purchased from Macklin biochemical technology Co., Ltd (Shanghai, China). NaBH4 was purchased from Aladdin Reagent Co., Ltd (Shanghai, China). Ti3AlC2 (200 mesh, purity >99%) was purchased from Beijing Forsman Scientific Co., Ltd (China). HF (48–51%) was purchased from Adamas Reagent Co., Ltd (Shanghai, China). Cuttlefish juice was purchased from bazaar (Qingdao, Shandong province, China). All reagents were used as received without any further treatment. Deionized (DI) water was used throughout the experiments.Instruments
The morphology, crystal structure and particle size of the samples were observed and analyzed by a Talos F200S field-emission transmission electron microscopy (TEM) and SU8220 field-emission scanning electron microscopy (SEM). LabRAM HR800 laser ultraviolet Raman spectrometer was used to obtain the information of molecular structure with the excitation wavelength's laser of 632.8 nm (17 mW). Escalab 250Xi monochromatization and dual-anode X-ray source X-ray photoelectron spectroscopy (XPS) were used to analyze all elements on the sample surface except He and to provide semi-quantitative analysis of relative content of elements. X-ray powder diffraction (XRD) analysis was performed on a D8 Advance diffractometer using Cu Ka radiation. Bruker A300 Electron Paramagnetic Resonance (EPR) with DMPO as a capture agent for hydroxyl radicals to explore mechanism. The PNP concentration in reaction solution was analyzed on UV-vis spectrometer (Lambda 25, PerkinElmer).Synthesis of catalyst
CQDs and MXene were prepared according to our previous work.32,33 Briefly, CQDs was prepared by a one-step hydrothermal method (180 °C, 12 h) of cuttlefish ink powder and MXene was produced by HF-etching (60 °C, 24 h) of Ti3AlC2.CQDs (0.5 mg mL−1), AgNO3 (0.068 g), and MXene (4.5, 1.0, 0.5 or 0.1 × 10−2 g) were added into round-bottomed flask under stirring at 80 °C for 12 h. The obtained reactant was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 min, and collected as CQD@Ag/MXene. Besides, CQD@Ag and CQD/MXene were also synthesized following the same procedures as above except without the addition of MXene or AgNO3, respectively. Equivalent mass Ag NPs and MXene were mixed then ultrasoniced for 30 min to obtain Ag/MXene.
000 rpm at 4 °C for 10 min, and collected as CQD@Ag/MXene. Besides, CQD@Ag and CQD/MXene were also synthesized following the same procedures as above except without the addition of MXene or AgNO3, respectively. Equivalent mass Ag NPs and MXene were mixed then ultrasoniced for 30 min to obtain Ag/MXene.
Evaluation of catalytic performance
The performance of Ag NPs, CQDs, MXene, Ag/MXene, CQD@Ag and CQD@Ag/MXene in the catalytic reduction of PNP were compared in the presence of NaBH4. In a typical experiment, 1 mL DI water, 15 μL PNP solution (10 mM) and 1 mL NaBH4 solution (66 mM) were mixed for 6 min. The well-mixing solutions were introduced into a quartz cuvette followed by the addition of 1 μL of 4 mg mL−1 Ag NPs, CQDs, MXene, Ag/MXene, CQD@Ag or CQD@Ag/MXene to the mixture. The reduction process was monitored by absorption spectra via a UV-vis spectrometer within a scanning range of 250–500 nm. Repeatability experiment was implemented by fixing the amount of catalyst, and repeated 10 times under the same conditions.The adsorption capacity of the catalysts was also evaluated by an adsorption experiment. In a typical procedure, 2.4 mg catalysts were added into 1 mL PNP (10 mg mL−1) aqueous solution, and the mixture was kept for 1.5 h to establish an adsorption–desorption equilibrium. Then, the supernatant was taken to monitor the concentration of PNP at the wavelength of 400 nm. The adsorption rate was calculated according to the following formula:
| Adsorbed percentage (%) = (C0 − Ce)/C0 × 100% | (1) |
Results and discussion
Morphology and elemental analysis
The morphology and particle size distribution of raw materials and catalysts were examined. MXene has a layered morphology (Fig. S1a†) and CQDs are uniformly dispersed spheres (Fig. S1b†). CQD@Ag particles are homogeneously distributed and loaded on MXene surface with layered structure shown in SEM (Fig. 1a) and TEM (Fig. S1c†) images of CQD@Ag/MXene, while the SEM (Fig. 1b) and TEM (Fig. S1d†) images of CQD@Ag prove that CQD@Ag easily agglomerated together and its particle size is different from CQD@Ag/MXene. The HRTEM image in Fig. 1c suggests that the CQD@Ag/MXene sample has a high crystallinity as evidenced by the clear lattice fringes, the interplanar spacing of the CQD@Ag supported on MXene is 0.228 nm, while CQD@Ag lattice fringes (Fig. 1d) is0.231 nm. Both of them are corresponding to the (111) lattice plane of Ag.34 The particle distribution of CQD@Ag on MXene is shown in the inset of Fig. S1c.† The particle size of CQD@Ag/MXene is in the range of 15.90–30.80 nm, and the average diameter is 22.80 nm which is smaller than that CQD@Ag (25.60 nm, Fig. S1d,† inset), indicating that the 2D material MXene can be used as an excellent support for uniform dispersion of CQD@Ag, which can effectively prevent the agglomeration of the CQD@Ag complex.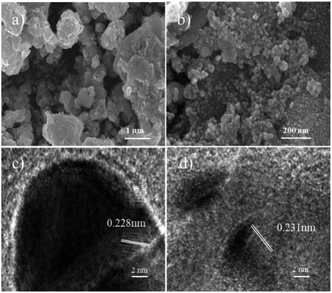 | ||
| Fig. 1 SEM images of (a) CQD@Ag/MXene and (b) CQD@Ag; HRTEM images of (c) CQD@Ag/MXene and (d) CQD@Ag. | ||
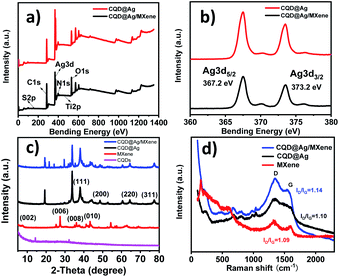
XPS was carried out to further investigate the composition and chemical status of the elements of CQD@Ag and CQD@Ag/MXene (Fig. 2a and b). Five main elements of C, N, O, S and Ag can be found (Fig. 2a). The relative content of each element is 68.63%, 20.20%, 4.36%, 0.25%, 6.20% in CQD@Ag/MXene and 67.78%, 18.47%, 4.45%, 0.39%, 8.91% in CQD@Ag, respectively. It is found that the relative content of Ag in CQD@Ag/MXene (6.20%) is lower than that in CQD@Ag (8.91%) because MXene was introduced and reduced CQD@Ag reunions. It's worth noting that CQD@Ag/MXene contains trace amount of Ti (0.37%). The peak at 454.60 eV is attributed to Ti2p. The peaks at around 284.8, 531.9, 399.4, 368.1 and 163.5 eV are attributed to C1s, O1s, N1s, Ag3d and S2p. Ag 3d5/2 and Ag 3d3/2 peaks in Fig. 2b are located at ∼367.2 eV and ∼373.2 eV. The slitting of the 3d doublet of Ag is 6.0 eV, which is in excellent agreement with the theoretical value of metallic Ag, indicating that the silver species exist as Ag0 on the surface of CQD@Ag and CQD@Ag/MXene.35 C–C, C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds can be observed in C1s spectrum (Fig. S2a and b†) both in CQD@Ag and CQD@Ag/MXene.36 The high-resolution O1s XPS spectrum (Fig. S2c and d†) of CQD@Ag and CQD@Ag/MXene is deconvoluted into four peaks at ∼530.7, ∼531.9, ∼533.3 and ∼534.6 eV which are attributed to the structures of Ag⋯O–, C–O–C, O
O bonds can be observed in C1s spectrum (Fig. S2a and b†) both in CQD@Ag and CQD@Ag/MXene.36 The high-resolution O1s XPS spectrum (Fig. S2c and d†) of CQD@Ag and CQD@Ag/MXene is deconvoluted into four peaks at ∼530.7, ∼531.9, ∼533.3 and ∼534.6 eV which are attributed to the structures of Ag⋯O–, C–O–C, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O and C–O–H.37 As shown in Fig. S2e and f,† the high-resolution N1s XPS spectra show that five major peaks of pyridine-N (398.3 eV), N–H (399.4 eV), pyrrole-N (400.3 eV), graphitic-N (401.1 eV) and Ag⋯N
C–O and C–O–H.37 As shown in Fig. S2e and f,† the high-resolution N1s XPS spectra show that five major peaks of pyridine-N (398.3 eV), N–H (399.4 eV), pyrrole-N (400.3 eV), graphitic-N (401.1 eV) and Ag⋯N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) (402.8 eV) can be observed on CQD@Ag/MXene, while only three peaks of pyridine-N (398.2 eV), N–H (399.4 eV) and Ag⋯N
(402.8 eV) can be observed on CQD@Ag/MXene, while only three peaks of pyridine-N (398.2 eV), N–H (399.4 eV) and Ag⋯N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) = (402.8 eV) on CQD@Ag.38
= (402.8 eV) on CQD@Ag.38
To gain more insights into the chemical environment and surface elemental composition, the CQD@Ag and CQD@Ag/MXene were further characterized by XRD and Raman spectrum. Seven diffraction peaks at 2θ value of 27°, 36°, 38°, 44°, 43°, 65°, 78° can be indexed to the (006), (008), (111), (200), (010), (220), (311) in the XRD pattern of CQD@Ag/MXene (Fig. 2c), which are the characteristic diffraction peaks of the XRD of CQD@Ag or MXene.32 It is obvious that diffraction peaks (111), (200), (220), and (311) in CQD@Ag/MXene attest the crystal faces of Ag,39–41 indicating the formation of crystalline Ag on the surface of CQDs.
Raman spectra (Fig. 2d) show that MXene has a strong peak at 150 cm−1 and three additional peaks at 394, 513 and 635 cm−1 which could be attributed to the vibrational modes of anatase.42 Two characteristic Raman peaks at 1346 and ∼1596 cm−1 can be observed on both composites and MXene, characterized as D band (defects and disorder) and G band (graphite), respectively. By comparison of the results, CQD@Ag/MXene shows a slight down-shift of G band, further confirming the accelerated electron transfer between MXene and CQD@Ag.43 The intensity ratio of D band to G band (ID/IG) is proportional to the amount of defect sites in carbon matrix, but relates to the degree of graphitization in inverse proportion.44 The higher ID/IG value (1.14) of CQD@Ag/MXene than 1.1 of CQD@Ag indicates that there are more defect sites in carbon matrix in CQD@Ag/MXene. The presence of more defect sites in carbon matrix facilitates the anchor and dispersion of active CQD@Ag, which may play a positive role in the catalytic reaction.38
Reduction of p-nitrophenol
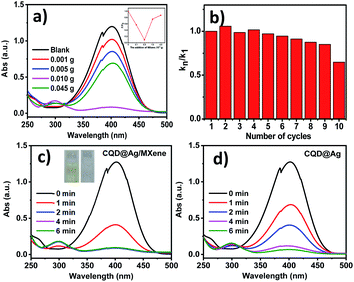
The reduction of PNP to PAP with NaBH4 was carried out to assess the catalytic performance of the CQD@Ag/MXene and CQD@Ag. Before that, the effect of different contents of MXene on the CQD@Ag/MXene was investigated. The pale yellow PNP solution turned dark yellow and had a maximum ultraviolet absorption peak at 400 nm after adding NaBH4 solution, which indicated the formation of p-nitrophenolate ions in the solution.45 The catalysts were then added to the mixed solution of PNP and NaBH4. As shown in Fig. 3a, the peak at 400 nm decreases in different degrees, and a new PAP peak appears at 300 nm under the catalysis of CQD@Ag/MXene,9 which indicates that the excessive or traces MXene content has different effect on the catalytic reduction rate of PNP. It can be clearly observed that reaction rate grows as the augment content of MXene, and the highest reaction rate reached when 0.005 g of MXene was used. Noted that, the catalytic reaction rate significantly decreases when the amount of MXene is more than 0.005 g. It is inferred that MXene itself has little catalytic activity for the reduction of PNP, superfluous MXene hinders the active site of CQD@Ag which can react with PNP. The reactive substance of CQD@Ag becomes less exposed in the unit MXene. Besides, a slight amount of MXene cannot load CQD@Ag completely, which resulted in the aggregation of CQD@Ag and masking of the active site. Thus 0.005 g of MXene is considered as the best dosage of the synthesis of CQD@Ag/MXene for the reaction.The stability and reusability of CQD@Ag/MXene were examined and results are shown in Fig. 3b. The reaction rate of the reduction PNP slightly decreases during the first ninth cycles. About 65% of the first cycle rate can be kept by the tenth cycle. The slightly decreasing of the reaction rate may be attributed to the accumulation of the reduced product PAP at the active sites of the CQD@Ag/MXene.46 The results show that CQD@Ag/MXene is a highly efficient catalyst with excellent stability and reusability.
As shown in Fig. 3c and d, the reaction solution became colorless and transparent (Fig. 3c, inset) about 2 min with CQD@Ag/MXene as the catalyst, which means that PNP is almost completely reduced to PAP. CQD@Ag/MXene exhibits superior catalysis reduction ability of PNP to CQD@Ag which took much longer to react (about 6 min) under the same conditions. The peak at 400 nm hardly decreases and no new absorption peak appears at 300 nm even for 12 min with single Ag NPs, CQDs and MXene as the catalyst (Fig. S3†). Ag/MXene (Fig. S3d†) and CQD/MXene (figure not given) just can reduce the peak intensity at 400 nm by 18.6% and 9.1% at 12 min which is much lower than CQD@Ag/MXene (91.3%) and CQD@Ag (68.2%) at 2 min. It indicates that Ag NPs, CQDs, MXene, Ag/MXene and CQD/MXene can hardly catalyze the reaction under the same catalytic conditions as CQD@Ag/MXene and CQD@Ag.
Catalytic kinetics and mechanism
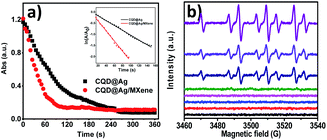
The pseudo first-order reaction kinetics can be applied to evaluate the catalytic rate because the concentration of BH4− remains constant during the reaction in the presence of excess NaBH4.47 As shown in Fig. 4a, the reduction reaction is rapid and the slope of the natural logarithmic plot of UV absorbance at 400 nm is the reaction rate constant k. Kinetics of PNP reduction in excess NaBH4 can be expressed by the linear eqn (2):45,48,49| ln(A/A0) = kt | (2) |
It can be deduced that the k for the reduction of PNP by CQD@Ag/MXene and CQD@Ag are 2.28 × 10−2 s−1 and 1.09 × 10−2 s−1, respectively. However, k is independent of the amount of catalysts. To objectively compare the catalytic activity of CQD@Ag/MXene and CQD@Ag with some other catalysts for PNP reduction reported in the literatures, the mass-normalized rate constants (km, km = k/m, where m denotes the mass of the catalyst) of some catalysts reported recently were calculated and tabulated in Table 1. It can be known that CQD@Ag/MXene and CQD@Ag show excellent catalytic performance. The km of CQD@Ag/MXene (5700 s−1 g−1) and CQD@Ag (2725 s−1 g−1) are superior to most of the noble metal-based catalysts in tens or even thousands times. Besides, due to the induction of Mxene which is a 2D material, the km of CQD@Ag/MXene is about twice that of CQD@Ag, indicating that Mxene has an indelible effect in increasing the performance of the catalyst.
| Catalyst | Catalyst amount (mg) | Reaction time (min) | Rate constant, k (s−1) | km (s−1 g−1) | Ref. |
|---|---|---|---|---|---|
| Ag/CuO | 1.0 | 6.0 | 0.00817 | 8.170 | 52 |
| Au/CuO | 1.0 | 8.0 | 0.00583 | 5.830 | 52 |
| Ni@Cu@Pd NPs | 0.010 | 3.0 | 0.00183 | 1830 | 53 |
| Cu/CuxO@C | 0.012 | 3.5 | 0.00110 | 916.7 | 38 |
| Cu/CuxO@CN | 0.012 | 0.7 | 0.12600 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
38 |
| Ag–COP | 3.0 | 5.0 | 0.01220 | 4.067 | 54 |
| Ag/ZrGP NPs(S1) | 10 | 16 | 0.00160 | 0.160 | 47 |
| Ag/ZrGP NPs(S2) | 10 | 10 | 0.00292 | 0.292 | 47 |
| Ag0@CZ–TEB | 2.0 | 2.0 | 0.01990 | 9.950 | 55 |
| Ag@TPHH-COF | 4.0 | 3.0 | 0.02200 | 5.500 | 37 |
| Ni/graphene nanostructure | 3.0 | 4.0 | 0.01170 | 3.900 | 56 |
| CuFeCN | 0.0018 | 4.0 | 0.01410 | 8057 | 57 |
| CQD@Ag | 0.0040 | 6.0 | 0.01090 | 2725 | This work |
| CQD@Ag/MXene | 0.0040 | 2.0 | 0.02280 | 5700 | This work |
The reaction pathway for the catalytic reduction of PNP to PAP has two steps. Firstly, the generated active hydrogen species which were converted by NaBH4 via hydrolysis reaction and PNP is adsorbed on catalyst surface. Next, the active hydrogen species will further react with PNP to PAP.50
The adsorption of PNP on the surface of the catalyst is the first step for its reduction reaction according to the Langmuir–Hinshelwood kinetics of heterogeneous catalysis.8 The adsorption ability of CQD@Ag and CQD@Ag/MXene to PNP is a favorable effect because the pre-adsorption of catalysts to PNP will significantly shorten the diffusion time of PNP to surface active sites of catalysts. Adsorption experiment was conducted to explore kinetics and mechanism. As shown in Fig. S4,† the ability of CQD@Ag/MXene to adsorb PNP is three times that of CQD@Ag, which benefits from MXene's layered structure and excellent adsorption properties. It may be one of the reasons why CQD@Ag/MXene has a faster catalytic speed than CQD@Ag.
Moreover, splendid catalyst can efficiently transfer electrons via active hydrogen species and react with PNP to form PAP. To make it clear, ESR was performed at room temperature using DMPO as a spin trap. As shown in Fig. 4b, when DMPO was added to a solution containing NaBH4, PNP and CQD@Ag/MXene, a high intensity EPR spectrum of 3460–3540 G was observed. The obtained spectrum consists of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 triplet of 1
1 triplet of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 triplets with aN = 16.6 G and aH = 22.5 G, which indicates the generation of large amount of H radical adducts.51 No H radical adducts can be detected in the solution of CQD@Ag/MXene or PNP with and without DMPO. However, H radical adducts were formed when PNP, NaBH4 and DMPO or NaBH4 and DMPO coexisted, which indicates that the H radical adduct was mainly derived from NaBH4. What's more, the strength of the H radical adduct was significantly enhanced when CQD@Ag/MXene was added. Indicating that CQD@Ag/MXene can promote the formation of H radicals in the solution, thereby increasing the reaction rate. CQD@Ag uniformly loaded on MXene has a larger specific surface area, which makes it easier for the substrate to be close to the reactive site to rapidly transfer electrons and H radicals, accelerating the production of PAP.
1 triplets with aN = 16.6 G and aH = 22.5 G, which indicates the generation of large amount of H radical adducts.51 No H radical adducts can be detected in the solution of CQD@Ag/MXene or PNP with and without DMPO. However, H radical adducts were formed when PNP, NaBH4 and DMPO or NaBH4 and DMPO coexisted, which indicates that the H radical adduct was mainly derived from NaBH4. What's more, the strength of the H radical adduct was significantly enhanced when CQD@Ag/MXene was added. Indicating that CQD@Ag/MXene can promote the formation of H radicals in the solution, thereby increasing the reaction rate. CQD@Ag uniformly loaded on MXene has a larger specific surface area, which makes it easier for the substrate to be close to the reactive site to rapidly transfer electrons and H radicals, accelerating the production of PAP.
As a consequence, the reaction kinetics and mechanism of the reduction of PNP with the CQD@Ag/MXene can be proposed as shown in Scheme 1.
Conclusion
We have developed two new kind composites of CQD@Ag/MXene and CQD@Ag. Both of them have the catalytic activity for the reduction of PNP with the km reaching up to 5700 and 2725 s−1 g−1, respectively. CQD@Ag/MXene exhibits better catalytic performance than CQD@Ag, probably due to the addition of MXene, which alleviates the agglomeration of CQD@Ag and exposes more active sites. The excellent catalytic activity of CQD@Ag/MXene may be attributed to its efficient adsorption of PNP and promotion of forming H radicals. CQD@Ag/MXene is promising as a highly effective catalyst for eliminating toxic chemicals in the environment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21876032).Notes and references
- R. Lamba, A. Umar, S. K. Mehta and S. K. Kansal, Talanta, 2015, 131, 490–498 CrossRef CAS PubMed.
- P. S. Da Silva, B. C. Gasparini, H. A. Magosso and A. Spinelli, J. Hazard. Mater., 2014, 273, 70–77 CrossRef PubMed.
- W. Liu, K. Tian, H. Jiang and H. Yu, Green Chem., 2014, 16, 4198–4205 RSC.
- G. Chang, Y. Luo, W. Lu, X. Qin, A. M. Asiri, A. O. Al-Youbi and X. Sun, Catal. Sci. Technol., 2012, 2, 800–806 RSC.
- W. Zhang, G. Li, W. Wang, Y. Qin, T. An, X. Xiao and W. Choi, Appl. Catal., B, 2018, 232, 11–18 CrossRef CAS.
- S. Singh, N. Kumar, M. Kumar, Jyoti, A. Agarwal and B. Mizaikoff, Chem. Eng. J., 2017, 313, 283–292 CrossRef CAS.
- E. Marais and T. Nyokong, J. Hazard. Mater., 2008, 152, 293–301 CrossRef CAS PubMed.
- T. B. Nguyen, C. P. Huang and R. Doong, Appl. Catal., B, 2019, 240, 337–347 CrossRef CAS.
- S. Saha, A. Pal, S. Kundu, S. Basu and T. Pal, Langmuir, 2010, 26, 2885–2893 CrossRef CAS PubMed.
- Y. Liu, Y. Y. Zhang, Q. W. Kou, D. D. Wang, D. L. Han, Z. Y. Lu, Y. Chen, L. Chen, Y. X. Wang, Y. J. Zhang, J. H. Yang and S. Xing, Powder Technol., 2018, 338, 26–35 CrossRef CAS.
- Y. Gao, J. Fang, Y. Zhang, C. Zhang, S. Zhao, Y. Zhou, M. Huang and X. Sheng, Appl. Organomet. Chem., 2018, 32, e4208 CrossRef.
- X. Li, L. Zhao, C. Shao, X. Li, W. Sun and Y. Liu, J. Colloid Interface Sci., 2018, 530, 345–352 CrossRef CAS PubMed.
- G. Liao, W. Zhao, Q. Li, Q. Pang and Z. Xu, Chem. Lett., 2017, 46, 1631–1634 CrossRef CAS.
- L. Zhang, X. Liu, Y. Wang and S. Xing, J. Alloys Compd., 2017, 709, 431–437 CrossRef CAS.
- S. M. Alshehri, T. Almuqati, N. Almuqati, E. Al-Farraj, N. Alhokbany and T. Ahamad, Carbohydr. Polym., 2016, 151, 135–143 CrossRef CAS PubMed.
- H. Cao, H. Huang, Z. Chen, B. Karadeniz, J. Lu and R. Cao, ACS Appl. Mater. Interfaces, 2017, 9, 5231–5236 CrossRef CAS PubMed.
- J. Zhang and L. Dai, ACS Catal., 2015, 5, 7244–7253 CrossRef CAS.
- Y. Cao, S. Mao, M. Li, Y. Chen and Y. Wang, ACS Catal., 2017, 7, 8090–8112 CrossRef CAS.
- H. Yu, H. Zhang, H. Huang, Y. Liu, H. Li, H. Ming and Z. Kang, New J. Chem., 2012, 36, 1031 RSC.
- S. Ghosh, S. S. Satapathy, K. Ghosh, S. Jauhari, S. K. Panda and S. Si, ChemistrySelect, 2019, 4, 3416–3422 CrossRef CAS.
- J. Liu, J. Li, R. Meng, P. Jian and L. Wang, J. Colloid Interface Sci., 2019, 551, 261–269 CrossRef CAS PubMed.
- B. Shao, Z. Liu, G. Zeng, H. Wang, Q. Liang, Q. He, M. Cheng, C. Zhou, L. Jiang and B. Song, J. Mater. Chem. A, 2020, 8, 758–7535 RSC.
- K. Wang, Y. Zhou, W. Xu, D. Huang, Z. Wang and M. Hong, Ceram. Int., 2016, 42, 8419–8424 CrossRef CAS.
- X. Guo, X. Xie, S. Choi, Y. Zhao, H. Liu, C. Wang, S. Chang and G. Wang, J. Mater. Chem. A, 2017, 5, 12445–12452 RSC.
- K. Rasool, M. Helal, A. Ali, C. E. Ren, Y. Gogotsi and K. A. Mahmoud, ACS Nano, 2016, 10, 3674–3684 CrossRef CAS PubMed.
- L. Ding, Y. Wei, Y. Wang, H. Chen, J. Caro and H. Wang, Angew. Chem., Int. Ed., 2017, 56, 1825–1829 CrossRef CAS PubMed.
- Y. Gao, L. Wang, Z. Li, A. Zhou, Q. Hu and X. Cao, Solid State Sci., 2014, 35, 62–65 CrossRef CAS.
- O. Mashtalir, K. M. Cook, V. N. Mochalin, M. Crowe, M. W. Barsoum and Y. Gogotsi, J. Mater. Chem. A, 2014, 2, 14334–14338 RSC.
- Y. Gao, L. Wang, A. Zhou, Z. Li, J. Chen, H. Bala, Q. Hu and X. Cao, Mater. Lett., 2015, 150, 62–64 CrossRef CAS.
- Y. Zhuang, Y. Liu and X. Meng, Appl. Surf. Sci., 2019, 496, 143647 CrossRef CAS.
- J. Zhang, Y. Zhao, X. Guo, C. Chen, C. Dong, R. Liu, C. Han, Y. Li, Y. Gogotsi and G. Wang, Nat. Catal., 2018, 1, 985–992 CrossRef CAS.
- Y. Wang, L. Zhou, X. Luo, Y. Zhang, J. Sun, X. A. Ning and Y. Yuan, ChemSusChem, 2018, 11, 4071–4076 CrossRef CAS PubMed.
- X. Huang, C. Yang, Y. Chen, Z. Zhu and L. Zhou, Anal. Methods, 2021, 13, 5351–5359 RSC.
- J. Feng, D. Fan, Q. Wang, L. Ma, W. Wei, J. Xie and J. Zhu, Colloids Surf., A, 2017, 520, 743–756 CrossRef CAS.
- M. Rakibuddin and R. Ananthakrishnan, New J. Chem., 2016, 40, 3385–3394 RSC.
- S. Chen, X. Hai, X. Chen and J. Wang, Anal. Chem., 2014, 86, 6689–6694 CrossRef CAS PubMed.
- R. Wang, D. Li, L. Wang, X. Zhang, Z. Zhou, J. Mu and Z. Su, Dalton Trans., 2019, 48, 1051–1059 RSC.
- W. Jia, F. Tian, M. Zhang, X. Li, S. Ye, Y. Ma, W. Wang, Y. Zhang, C. Meng, G. Zeng and J. Liu, J. Colloid Interface Sci., 2021, 594, 254–264 CrossRef CAS PubMed.
- F. Zhang, K. Zhang, F. Xie, J. Liu, H. Dong, W. Zhao and Z. Meng, Appl. Surf. Sci., 2013, 265, 578–584 CrossRef CAS.
- W. Yuan, Y. Gu and L. Li, Appl. Surf. Sci., 2012, 261, 753–758 CrossRef CAS.
- Y. Jiang, X. Zhang, L. Pei, S. Yue, L. Ma, L. Zhou, Z. Huang, Y. He and J. Gao, Chem. Eng. J., 2018, 339, 547–556 CrossRef CAS.
- Z. Xu, Y. Sun, Y. Zhuang, W. Jing, H. Ye and Z. Cui, J. Membr. Sci., 2018, 564, 35–43 CrossRef CAS.
- G. Zuo, Y. Wang, W. L. Teo, A. Xie, Y. Guo, Y. Dai, W. Zhou, D. Jana, Q. Xian, W. Dong and Y. Zhao, Chem. Eng. J., 2021, 403, 126328 CrossRef CAS.
- Z. Hasan, D. Cho, C. Chon, K. Yoon and H. Song, Chem. Eng. J., 2016, 298, 183–190 CrossRef CAS.
- P. Veerakumar, R. Madhu, S. Chen, V. Veeramani, C. Hung, P. Tang, C. Wang and S. Liu, J. Mater. Chem. A, 2014, 2, 16015–16022 RSC.
- F. Lin and R. Doong, J. Phys. Chem. C, 2011, 115, 6591–6598 CrossRef CAS.
- A. Zhou, J. Li, G. Wang and Q. Xu, Appl. Surf. Sci., 2020, 506, 144570 CrossRef CAS.
- Y. Peng, X. Wu, L. Qiu, C. Liu, S. Wang and F. Yan, J. Mater. Chem. A, 2013, 1, 9257–9263 RSC.
- L. Ai, H. Yue and J. Jiang, J. Mater. Chem., 2012, 22, 23447–23453 RSC.
- G. Liao, Y. Gong, L. Zhong, J. Fang, L. Zhang, Z. Xu, H. Gao and B. Fang, Nano Res., 2019, 12, 2407–2436 CrossRef CAS.
- T. Aditya, J. Jana, N. K. Singh, A. Pal and T. Pal, ACS Omega, 2017, 2, 1968–1984 CrossRef CAS PubMed.
- E. Akbarzadeh, M. Falamarzi and M. R. Gholami, Mater. Chem. Phys., 2017, 198, 374–379 CrossRef CAS.
- Z. Gong, T. Ma and F. Liang, J. Alloys Compd., 2021, 873, 159802 CrossRef CAS.
- F. Rezaei and M. Dinari, Colloids Surf., A, 2021, 618, 126441 CrossRef CAS.
- W. Gong, Q. Wu, G. Jiang and G. Li, J. Mater. Chem. A, 2019, 7, 13449–13454 RSC.
- Y. Wu, M. Wen, Q. Wu and H. Fang, J. Phys. Chem. C, 2014, 118, 6307–6313 CrossRef CAS.
- T. Wi-Afedzi, E. Kwon, D. D. Tuan, K. A. Lin and F. Ghanbari, Sci. Total Environ., 2020, 703, 134781 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra09177h |
| This journal is © The Royal Society of Chemistry 2022 |