Open Access Article
Open Access ArticleGeneration of novel Diels–Alder reactions using a generative adversarial network†
Sheng Li‡
 ab,
Xinqiao Wang‡b,
Yejian Wu
ab,
Xinqiao Wang‡b,
Yejian Wu b,
Hongliang Duanbc and
Lan Tang*a
b,
Hongliang Duanbc and
Lan Tang*a
aCollege of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou 310014, P. R. China. E-mail: tanglan@zjut.edu.cn
bArtificial Intelligence Aided Drug Discovery Institute, College of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou 310014, P. R. China
cState Key Laboratory of Drug Research, Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences, Shanghai 201203, China
First published on 25th November 2022
Abstract
Deep learning has enormous potential in the chemical and pharmaceutical fields, and generative adversarial networks (GANs) in particular have exhibited remarkable performance in the field of molecular generation as generative models. However, their application in the field of organic chemistry has been limited; thus, in this study, we attempt to utilize a GAN as a generative model for the generation of Diels–Alder reactions. A MaskGAN model was trained with 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 Diels–Alder reactions, and 1441 novel Diels–Alder reactions were generated. Analysis of the generated reactions indicated that the model learned several reaction rules in-depth. Thus, the MaskGAN model can be used to generate organic reactions and aid chemists in the exploration of novel reactions.
092 Diels–Alder reactions, and 1441 novel Diels–Alder reactions were generated. Analysis of the generated reactions indicated that the model learned several reaction rules in-depth. Thus, the MaskGAN model can be used to generate organic reactions and aid chemists in the exploration of novel reactions.
Introduction
Organic chemistry has played a significant role in human history, and organic reactions have been vital in the synthesis of chemicals for the development of drugs and materials for hundreds of years. Prolonged experiment times, high experiment costs, and low success rates have hindered the exploration of conventional chemical reactions.1 Fortunately, the advent of artificial intelligence (AI) has provided researchers with novel strategies for the development of organic chemistry-related applications.2In recent years, owing to the constant development of computational techniques, AI has achieved remarkable results in the fields of retrosynthesis and reaction prediction.3,4 Zheng et al. applied a transformer model to develop a template-free self-corrected retrosynthesis predictor and predicted retrosynthesis reactions with an accuracy of 59.0% on a standard benchmark data set.5 Wang et al. reported a method that utilized transfer learning to enhance the accuracy of a transformer model (94.9%), which is higher than the accuracy of a transformer-baseline model (66.3%).6 Recently, the application of deep neural networks in the field of pharmaceutical chemistry, particularly for drug molecular generation and toxicity risk assessment, has received extensive attention.7,8 Lee et al. applied a generative adversarial network (GAN) to de novo molecular design and demonstrated high performance in the five distribution learning benchmarks of the GuacaMol framework.9 The success of generative models in molecular generation inspired researchers such as Bort et al. and Wang et al. to further explore reaction generative models.10,11
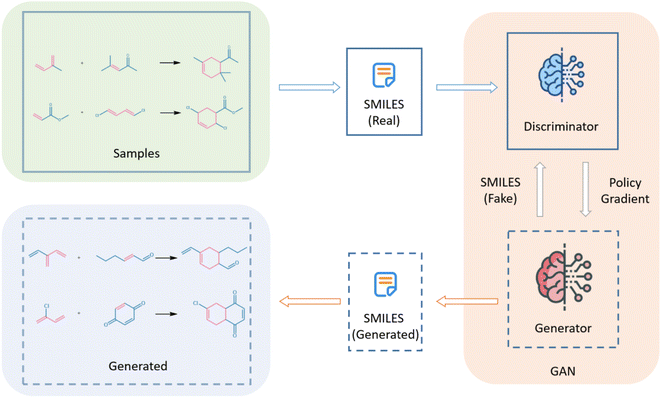
Generative models are an important class of machine learning models capable of generating new data that is not included in a training dataset and have exhibited considerable potential in image,12 text,13 and sound generation14 in the past few years. Among several generative deep learning models, GAN has attracted considerable attention from researchers and was first proposed by Goodfellow et al. as a novel framework for the estimation of generative models in adversarial processes.15 GAN utilizes two adversarial networks, a generator that captures the distribution of data and a discriminator that estimates the probability of a sample belonging to the training data. They compete until the discriminator is unable to distinguish between the real data and the data generated by the generator. This operating mechanism of GAN has allowed it to exhibit superiority over other models via more realistic image generation.16 However, the application of GAN in the field of chemistry had been limited towing to the discreteness of simplified molecular-input line-entry system (SMILES) strings, which replace molecular structures as the input data. To overcome this issue, Sutton et al. proposed a policy gradient-based reinforcement learning approach on GAN that provided feedback on information and has been applied by several scientists.17 Lin et al. used GANs for de novo molecular design, dimensionality reduction, and de novo peptide and protein design,18 while Maziarka et al. reported an improved cycleGAN-based molecular optimization model called mol-CycleGAN that could generate optimized compounds with the desired properties and structures similar to the originally provided molecules.19 Prykhodko et al. proposed a novel deep learning architecture called LatentGAN for de novo molecular design, that combined autoencoders and GANs.20 In this study, we attempt to utilize GANs for generating novel reactions and enhance the scope of chemical reactions. Fig. 1 shows a flowchart for the generation of Diels–Alder reactions using GAN, where “Samples” refers to the dataset and “Generated” refers to the novel reactions generated by the GAN. In this study, we converted the reactions of the dataset into SMILES strings prior to importing them into the GAN model. Subsequently, the discriminator and generator of the GAN were trained to generate the novel Diels–Alder reactions.
The dataset for the GAN model to learn has a significant effect on the model's performance, and thus an applicable dataset must be chosen. As the Diels–Alder reaction is one of the most effective and widely used organic reactions in drug and material synthesis, we chose the Diels–Alder reaction as the training dataset in this study. The Diels–Alder cycloaddition reaction consists of the cyclization of a diene and alkene to form a cyclohexene derivative and was discovered by O. Diels and K. Alder when they established the structure of the cycloadduct of p-quinone and cyclopentadiene in 1928.21 Since its discovery, extensive data on Diels–Alder reactions have been reported, which is another reason for choosing the Diels–Alder reaction dataset in this study. Boger et al. reported that a key step in the synthesis of rubrolone aglycon's seven-membered C-ring in a laboratory was the intermolecular Diels–Alder reaction of an electron-rich diene with an extremely strained dienophile. Furthermore, they reported an excellent cycloaddition yield of 97% with the products exhibiting complete enantioselectivity.22 In a Diels–Alder reaction, the reactants move closer to each other and interact to form a cyclic transition state, which gradually transforms into a product molecule.
In this study, we utilized MaskGAN, which is composed of a generator, discriminator, and critic network, for the generation of novel Diels–Alder reactions.23 The generator network uses a sequence-to-sequence model with an attention mechanism. The Reaxys database was used to construct a training dataset of Diels–Alder reactions, which were converted to SMILES strings and then imported to the MaskGAN model for training and reaction generation. The model generated 1441 novel Diels–Alder reactions and they were compared with the Diels–Alder reaction dataset to verify their novelty. Using the newly discovered reactions from this study combined with AI we could accelerate the discovery of reactions and consequently enhance the accuracy of the prediction of organic reactions.
Method
Dataset
To train MaskGAN for the generation of Diels–Alder reactions, a dataset of Diels–Alder reactions downloaded from the Reaxys database was created. The keyword “Diels–Alder Reaction” was used to find the reactions on the database, and duplicate reactions and invalid reactions (reactions with empty reactants or products and reactions with reactants equal to products) were deleted. An RDKit template was used to screen the chemical feasibility of the Diels–Alder reactions and finally, a dataset of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 reactions was assembled. The dataset was split into a training set and validation set in a ratio of 8
092 reactions was assembled. The dataset was split into a training set and validation set in a ratio of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.
2.
Model
Initially, GANs were designed to output differentiable values, and thus it is difficult for GANs to generate discrete language. To overcome this issue, MaskGAN, an actor-critic24,25 conditional GAN was introduced by filling in missing text conditioned to generate higher quality samples.The generator network of the MaskGAN uses a sequence-to-sequence architecture26 with an attention mechanism.27 The actual implementation of seq2seq in a MaskGAN is a form of a long short-term memory (LSTM) network,28 containing an encoder and a decoder structure. The encoder processes every element in the input sequence and compiles the captured information into a context vector. Then, the encoder sends the context vector to the decoder, which starts producing the output sequence item-by-item to eventually produce the entire sentence. The discriminator uses the same seq2seq structure as the generator with the exception that a scalar probability is output at every time step. In addition, for converging more rapidly, the critic in MaskGAN helps the generator by reducing the high-variance of the gradient updates in a high action-space environment, which enables a more stable training procedure.
To train the model, 80% of the downloaded Diels–Alder reactions were applied while the remainder were used for validation. The model was trained with a batch size of 512 for 300 epochs, the masking ratio of the input sequence was set to 0.1, and the network parameters were optimized using Adam optimization with a weight decay of 0.001. The base learning rate was set to 0.01 and was multiplied by 0.9 to reduce the learning rate for every epoch. All experiments were implemented using PyTorch 1.7.0 (for the detailed version of the package, please access the URL: https://github.com/hongliangduan/Generation-of-novel-Diels-Alder-reaction-using-a-GAN-.git).
Result and discussion
To generate novel reactions using our model, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 Diels–Alder reactions were imported from the Reaxys database into the model for generating reactions. After reaction generation, we removed generated reactions with invalid SMILES strings for reactants or products automatically by using RDkit software. However, the generated reaction may still not conform to the Diels–Alder reaction mechanism, although the SMILES strings of the reactants and products of all reactions were valid. Therefore, through screening of the generated reactions using reaction templates of the RDKit module, reactions that do not conform to the Diels–Alder reaction mechanism were discarded. However, owing to the limitations of the RDKit, we were unable to automatically remove reactions with chiral errors, and thus, these reactions had to be removed manually after the screening with the RDKit. In this study, 26
092 Diels–Alder reactions were imported from the Reaxys database into the model for generating reactions. After reaction generation, we removed generated reactions with invalid SMILES strings for reactants or products automatically by using RDkit software. However, the generated reaction may still not conform to the Diels–Alder reaction mechanism, although the SMILES strings of the reactants and products of all reactions were valid. Therefore, through screening of the generated reactions using reaction templates of the RDKit module, reactions that do not conform to the Diels–Alder reaction mechanism were discarded. However, owing to the limitations of the RDKit, we were unable to automatically remove reactions with chiral errors, and thus, these reactions had to be removed manually after the screening with the RDKit. In this study, 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 869 reactions were generated by MaskGAN, from which 13
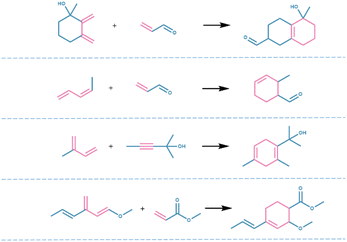
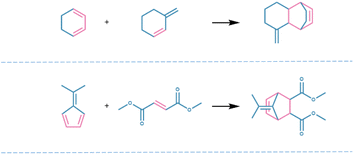
869 reactions were generated by MaskGAN, from which 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 reactions were removed automatically by the RDKit. Subsequently, the duplicate generated reactions and the generated reactions that also belonged to the training set were removed to obtain 1881 novel reactions, from which 441 reactions with chiral errors were removed manually, and eventually, 1441 novel Diels–Alder reactions were obtained from the model (Fig. 2). Fig. 3 shows the practical reactions representatively selected from the generated set.
320 reactions were removed automatically by the RDKit. Subsequently, the duplicate generated reactions and the generated reactions that also belonged to the training set were removed to obtain 1881 novel reactions, from which 441 reactions with chiral errors were removed manually, and eventually, 1441 novel Diels–Alder reactions were obtained from the model (Fig. 2). Fig. 3 shows the practical reactions representatively selected from the generated set.
To further investigate the generated novel reactions, they were analyzed at the molecular level. Table 1 shows the total amount of valid molecules and their proportion out of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generated molecules for every component in the reactions. Table 2 shows the amounts and proportions of reactants and products in the generated set calculated using different metrics.
000 generated molecules for every component in the reactions. Table 2 shows the amounts and proportions of reactants and products in the generated set calculated using different metrics.
| Components in the reactions | Valid molecular | ||
|---|---|---|---|
| Total | Amount | Rate | |
| Dienes | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7012 | 70.1% |
| Dienophiles | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7483 | 74.8% |
| Products | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
3048 | 30.5% |
| Components in the reactions | Unique molecular | Novel molecular | ||
|---|---|---|---|---|
| Amount | Rate | Amount | Rate | |
| Dienes | 661 | 42.8% | 452 | 68.3% |
| Dienophiles | 825 | 62.0% | 628 | 76.1% |
| Products | 1394 | 97.0% | 1035 | 74.2% |
Validity
The validity of the model was calculated as the ratio of the number of valid molecules to generated molecules. The validities with respect to dienes, dienophiles, and product components were 70.1%, 74.8%, and 30.5%, respectively. These values suggest that our model exhibits outstanding validity for dienes and dienophiles, whereas a moderate validity for the products indicates scope for optimization.Uniqueness
The uniqueness of the model was computed as the ratio of unique molecules in the generated set to valid molecules. The uniqueness of the model with respect to the products, dienes, and dienophiles components was 97.0%, 42.8%, and 62.0%, respectively. We speculate that the superior uniqueness of the products is owing to their complex structure. The high uniqueness ratios indicated that the model does not generate only a few typical molecules and is capable of generating a large number of reactions with unique products.Novelty
We counted the number of novel molecules of each component in the reactions and the novelty of the model was estimated as the ratio of the generated molecules not in the training set to the unique molecules in the generated set. The number of novel dienophiles was 628 while its proportion was 76.1%, which indicates that our model preferentially generated reactions containing novel dienophiles rather than replacing various reactants to generate products the same as those in the training set. The proportions of novel dienes and product components were 68.3% and 74.2%. Although the proportion of novel dienes is moderate, the ability of reactants to combine in pairs enables the possibility of generation of a large number of novel products without the need for a large number of novel reactants.As shown in Fig. 4, we observed that when three carbon–carbon double bonds were present in the reactants, the two double bonds of dienophile components chose to react in the s-cis conformation in 98.4% of the reactions in the generated set. This indicates that our model can effectively learn reaction mechanisms. During a Diels–Alder reaction, the s-cis conformation is more favorable in the formation of the transition state. Therefore, dienes that are permanently in the s-trans conformation and cannot adopt the s-cis conformation will not undergo the Diels–Alder reaction. The two ends of these dienes cannot get close enough to the dienophiles in Diels–Alder reactions and could thus result in the formation of a novel six-membered ring of products with a trans double bond. On the contrary, dienes that are permanently in the s-cis conformation, such as cyclic dienes, significantly favor Diels–Alder reactions.
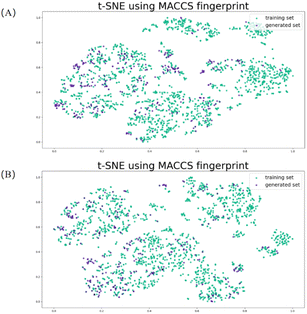
The generated reactions were further analyzed to establish a correlation between the generated reactions and the Diels–Alder reactions of the training set. The reactants were distributed using MACCS29 molecular fingerprints and the t-distributed stochastic neighbor embedding (t-SNE)30 method. t-SNE is a variation on the stochastic neighbor embedding method proposed by Maaten et al.31 and is easier to optimize and reduces the tendency of points to cluster in the center of the map.
MACCS fingerprints are a molecular qualitative descriptor comprising high-dimensional data of 166-dimensional molecular features of various functional groups and 1-dimensional placeholders. t-SNE was used as a dimensionality reduction technique to visualize the MACCS molecular fingerprints of the reactants. Fig. 5(A) shows the t-SNE plot of the distribution of the MACCS fingerprints of the novel dienophile components in generated set and the dienophiles in the training dataset. We observed that the distribution of the training set adequately covered the generated set, which indicates that while the dienophile components generated by the model are novel, they satisfy the features of the reactants of Diels–Alder reactions as well. A similar observation was made with respect to the diene components Fig. 5(B). These results effectively prove that the generated reactions follow the distribution of the features of the training dataset.
The generated reactions were further analyzed based on the level of chemical transformation. Table 3 summarizes the amounts and proportions of chemically feasible, unique, and novel reactions out of the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generated valid reactions. Table 4 shows the proportion of reactions that conform to the regioselectivity and stereospecificity of the Diels–Alder reaction. A chemically feasible reaction conforms to a particular reaction mechanism, and in this study, the reaction mechanism is that of a Diels–Alder reaction. A valid reaction is one in which the products and reactants of the reaction are chemically valid. The chemical feasibility of the model was computed as the ratio of the number of chemically feasible reactions that were screened by the RDKit templates to the number of valid reactions in the generated set. While the proportion of the chemically feasible generated reactions after excluding duplicate chemically feasible reactions indicates the uniqueness of the model, the proportion of the molecules that are present in the training dataset indicates its novelty. The chemical feasibility, uniqueness, and novelty of the model were estimated to be 50.4%, 40.6%, and 21.4%, respectively, which indicate that the model exhibits moderate novelty. We believe this is due to the limited chemical space constituted by the training set with small data. Given that pre-training and data augmentation can improve the training effect of small data. For future work, the utilization of pre-training and data augmentation methods can be explored to overcome this issue. In addition, of 10
000 generated valid reactions. Table 4 shows the proportion of reactions that conform to the regioselectivity and stereospecificity of the Diels–Alder reaction. A chemically feasible reaction conforms to a particular reaction mechanism, and in this study, the reaction mechanism is that of a Diels–Alder reaction. A valid reaction is one in which the products and reactants of the reaction are chemically valid. The chemical feasibility of the model was computed as the ratio of the number of chemically feasible reactions that were screened by the RDKit templates to the number of valid reactions in the generated set. While the proportion of the chemically feasible generated reactions after excluding duplicate chemically feasible reactions indicates the uniqueness of the model, the proportion of the molecules that are present in the training dataset indicates its novelty. The chemical feasibility, uniqueness, and novelty of the model were estimated to be 50.4%, 40.6%, and 21.4%, respectively, which indicate that the model exhibits moderate novelty. We believe this is due to the limited chemical space constituted by the training set with small data. Given that pre-training and data augmentation can improve the training effect of small data. For future work, the utilization of pre-training and data augmentation methods can be explored to overcome this issue. In addition, of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reactions generated with MaskGAN, 438 turn out to be chemically meaningful and novel. Therefore, our success rate is 4.38%. Wang et al. describe a Transformer-Based reaction generation strategy, and their success rate is 2.86% after the same data processing, our method has about 1.5% improvement.11
000 reactions generated with MaskGAN, 438 turn out to be chemically meaningful and novel. Therefore, our success rate is 4.38%. Wang et al. describe a Transformer-Based reaction generation strategy, and their success rate is 2.86% after the same data processing, our method has about 1.5% improvement.11
| Total generated valid reactions | Chemically feasible reactions | Unique reactions | Novel reactions | |||
|---|---|---|---|---|---|---|
| Amount | Rate | Amount | Rate | Amount | Rate | |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
5042 | 50.4% | 2047 | 40.6% | 438 | 21.4% |
| Rule | Rate |
|---|---|
| Regioselectivity | 100% |
| Stereospecificity | 76.6% |
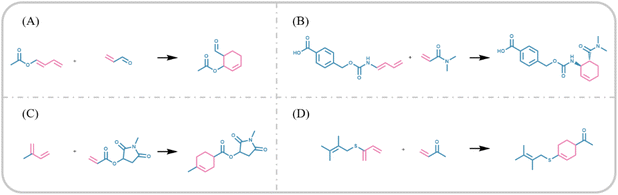
As shown in Table 4, we observed a 100% probability that the model generates reactions with an ortho or para product, which is consistent with our knowledge of the regioselectivity of Diels–Alder reactions, indicating that our model exhibits excellent regioselectivity. Regioselectivity refers to the preference of a reagent to react with a functional group at a particular position over another. Diels–Alder reactions are highly regioselective as one of the carbon–carbon double bonds in a diene is more likely to be attacked by a dienophile at its electrophilic site rather than its electron-donating group. As shown in Fig. 6(A) and (B), the presence of the electron-donating group at one end of the diene results in the other end being more electrophilic, resulting in a higher preference for dienophile components to attack the electrophilic site. When the electron-donating group is located at an end of the diene, the dienophile tends to attack the other end of the diene to produce ortho products, whereas when the electron-donating group is in the middle of the diene (Fig. 6(C) and (D)), the dienophile attack the carbon–carbon double bond of the electron-donating group to produce para products. Therefore, the Diels–Alder reaction is a cycloaddition reaction consisting of an aromatic transition state that is ortho and para directing, and the reactions generated by our model conform to this mechanism.
Stereoselectivity refers to the preferential reaction of a reactant based on the stereochemistry of the product. The mechanism of product formation in Diels–Alder reaction follows the endo rule in which the electron-withdrawing group of the dienophile components and the newly formed carbon–carbon double bond in the middle of the old diene tend to be on the same side during the process, forming an endo product. The bonding interaction between the electron-withdrawing group of the dienophile and the π bond formed at the back of the diene result in an increased rate of endo product formation. In irreversible Diels–Alder reactions, endo products are preferred as kinetic products, whereas in reversible Diels–Alder reactions, exo products are formed instead as exo products are more stable than endo products owing to their lesser steric hindrance. As only irreversible Diels–Alder reactions were selected for the training set, the majority of the generated reactions contain only endo products. The stereoselectivity of one of the novel reactions generated by our model is depicted in Fig. 7(A). The asymmetric dienophile reacts with the cyclic diene, resulting in the formation of carbonyl groups on the dienophile and the newly formed double bond in the middle of the old diene on the same side and hydrogen atoms above the generated ring. This product is an endo product, which conforms to the stereoselectivity rule of Diels–Alder reactions and indicates that our model learned the stereoselectivity of Diels–Alder reactions.
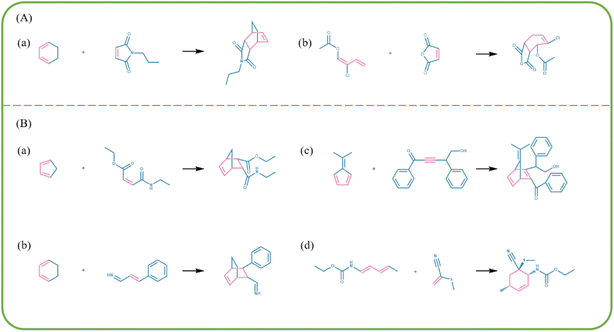 | ||
| Fig. 7 Analysis of the generated reactions. (A) Stereoselectivity of the generated reactions. (B) Stereospecificity of the generated reactions. | ||
On further analysis of the generated reactions, we observed that the structure of the products mostly depended on the structure of the reactants, which indicated the stereospecificity of the generated Diels–Alder reactions. As shown in Fig. 7(B)(a), the product sustains a cis configuration when the dienophile component with a cis configuration reacts with the diene in the Diels–Alder reaction, and similarly, the trans configuration is sustained from the dienophile reactant to the product (Fig. 7(B)(b)). During the transition state of the reaction of dienophiles with a trans configuration, one of the functional groups gets tucked under the diene and then reappears underneath the ring when the product molecule is formed to reproduce the trans configuration. The configuration of the diene components Diels–Alder reactions also exhibit a significant influence on the configuration of the products. Fig. 7(B)(c) shows that as both carbon–carbon double bonds of the diene are in cis conformation, the two hydrogen atoms are present below the newly formed six-membered ring. Whereas, when the two carbon–carbon double bonds are in trans configuration, the functional groups of the dienes lie outside the newly formed six-membered ring Fig. 7(B)(d). Therefore, the products of the generated reactions effectively reproduced the stereochemistry of the dienophiles and as shown in Table 4, 76.6% of the reactions with products exhibiting cis/trans isomerism exhibited stereospecificity, which indicated that Diels–Alder reactions are mostly stereospecific.
Conclusion
In this study, we trained the MaskGAN model with a dataset containing 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 Diels–Alder reactions and consequently generated 1441 novel Diels–Alder reactions. To establish a correlation between the generated and training dataset reactions, the generated novel reactions were artificially judged using the Diels–Alder reaction templates from RDKit package. On analysis of the validity, uniqueness and novelty of reaction components and reaction mechanism of the generated reactions, we concluded that the reactions satisfied the majority of the features of Diels–Alder reactions. Our model exhibited excellent performance with respect to regioselectivity and the rule that dienes with the s-cis conformation are considerably favorable for Diels–Alder reactions, which indicated that MaskGAN exhibited a clear understanding of the intrinsic rules of Diels–Alder reactions. The objective of our study was to generate novel reactions that conform to mechanism via GAN to ultimately aid in the development of de novo reaction designs without additional import. The model reported in this study is no longer limited by imports and can thus provide chemists with ESI† on novel reactions and facilitate the exploration of novel reactions.
092 Diels–Alder reactions and consequently generated 1441 novel Diels–Alder reactions. To establish a correlation between the generated and training dataset reactions, the generated novel reactions were artificially judged using the Diels–Alder reaction templates from RDKit package. On analysis of the validity, uniqueness and novelty of reaction components and reaction mechanism of the generated reactions, we concluded that the reactions satisfied the majority of the features of Diels–Alder reactions. Our model exhibited excellent performance with respect to regioselectivity and the rule that dienes with the s-cis conformation are considerably favorable for Diels–Alder reactions, which indicated that MaskGAN exhibited a clear understanding of the intrinsic rules of Diels–Alder reactions. The objective of our study was to generate novel reactions that conform to mechanism via GAN to ultimately aid in the development of de novo reaction designs without additional import. The model reported in this study is no longer limited by imports and can thus provide chemists with ESI† on novel reactions and facilitate the exploration of novel reactions.
Code availability
The code and the trained model are available from https://github.com/hongliangduan/Generation-of-novel-Diels-Alder-reaction-using-a-GAN-.git.Data availability
The training and validation used in our study are available from https://github.com/hongliangduan/Generation-of-novel-Diels-Alder-reaction-using-a-GAN-/tree/main/dataset. Source data are provided with this paper.Author contributions
These authors contributed equally: S. L. and X. W., H. D. conceived the presented idea. S. L. trained models. S. L. and Y. W. analyzed the data. X. W. and S. L. wrote the manuscript. All authors discussed the results and approved the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This project was supported by the National Natural Science Foundation of China (No. 81903438), Natural Science Foundation of Zhejiang Province (LD22H300004) and the Public welfare project of Zhejiang Science and Technology Department [Grant No. GF20H280010].Notes and references
- A. Lavecchia, Drug Discovery Today, 2019, 24, 2017–2032 CrossRef PubMed.
- A. C. Mater and M. L. Coote, J. Chem. Inf. Model., 2019, 59, 2545–2559 CrossRef CAS PubMed.
- S. Ishida, K. Terayama, R. Kojima, K. Takasu and Y. Okuno, J. Chem. Inf. Model., 2019, 59, 5026–5033 CrossRef CAS PubMed.
- D. Fooshee, A. Mood, E. Gutman, M. Tavakoli, G. Urban, F. Liu, N. Huynh, D. Van Vranken and P. Baldi, Mol. Syst. Des. Eng., 2018, 3, 442–452 RSC.
- S. Zheng, J. Rao, Z. Zhang, J. Xu and Y. Yang, J. Chem. Inf. Model., 2019, 60, 47–55 CrossRef PubMed.
- L. Wang, C. Zhang, R. Bai, J. Li and H. Duan, Chem. Commun., 2020, 56, 9368–9371 RSC.
- H. Chen, O. Engkvist, Y. Wang, M. Olivecrona and T. Blaschke, Drug Discovery Today, 2018, 23, 1241–1250 CrossRef PubMed.
- W. Tang, J. Chen, Z. Wang, H. Xie and H. Hong, J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev., 2018, 36, 252–271 CrossRef CAS PubMed.
- Y. J. Lee, H. Kahng and S. B. Kim, Mol. Inf., 2021, 40, 2100045 CrossRef CAS PubMed.
- W. Bort, I. I. Baskin, T. Gimadiev, A. Mukanov, R. Nugmanov, P. Sidorov, G. Marcou, D. Horvath, O. Klimchuk and T. Madzhidov, Sci. Rep., 2021, 11, 1–15 CrossRef PubMed.
- X. Wang, C. Yao, Y. Zhang, J. Yu, H. Qiao, C. Zhang, Y. Wu, R. Bai and H. Duan, J. Cheminf., 2022, 14, 60 Search PubMed.
- A. Brock, J. Donahue and K. Simonyan, arXiv, 2018, preprint, arXiv:1809.11096, DOI:10.48550/arXiv.1809.11096.
- W. Nie, N. Narodytska and A. Patel, in ICLR, 2018 Search PubMed.
- S. Liu, S. Li and H. Cheng, IEEE Trans. Circuits Syst. Video Technol., 2021, 32, 1299–1312 Search PubMed.
- I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville and Y. Bengio, Adv. Neural Inf. Process Syst., 2014, 27, 2672–2680 Search PubMed.
- J. Gu, Y. Shen and B. Zhou, in CVPR. 2020 Search PubMed.
- R. S. Sutton, D. McAllester, S. Singh and Y. Mansour, Adv. Neural Inf. Process Syst., 1999, 12, 1057–1063 Search PubMed.
- E. Lin, C.-H. Lin and H.-Y. Lane, Molecules, 2020, 25, 3250 CrossRef CAS PubMed.
- Ł. Maziarka, A. Pocha, J. Kaczmarczyk, K. Rataj, T. Danel and M. Warchoł, J. Cheminf., 2020, 12, 1–18 Search PubMed.
- O. Prykhodko, S. V. Johansson, P.-C. Kotsias, J. Arús-Pous, E. J. Bjerrum, O. Engkvist and H. Chen, J. Cheminf., 2019, 11, 1–13 Search PubMed.
- O. Diels, Ber. Dtsch. Chem. Ges., 1929, 62, 554–562 CrossRef.
- D. L. Boger and J. S. Panek, J. Org. Chem., 1981, 46, 2179–2182 CrossRef CAS.
- W. Fedus, I. Goodfellow and A. M. Dai, arXiv, 2018, preprint, arXiv:1801.07736, DOI:10.48550/arXiv.1801.07736.
- V. Konda and J. Tsitsiklis, Adv. Neural Inf. Process Syst., 1999, 12, 1008–1014 Search PubMed.
- I. Grondman, L. Busoniu, G. A. Lopes and R. Babuska, IEEE Trans. Syst. Man Cybern. C: Appl. Rev., 2012, 42, 1291–1307 Search PubMed.
- I. Sutskever, O. Vinyals and Q. V. Le, Adv. Neural Inf. Process Syst., 2014, 27, 3104–3112 Search PubMed.
- M.-T. Luong, H. Pham and C. D. Manning, arXiv, 2015, preprint, arXiv:1508.04025, DOI:10.48550/arXiv.1508.04025.
- S. Hochreiter and J. Schmidhuber, Neural Comput., 1997, 9, 1735–1780 CrossRef CAS PubMed.
- J. L. Durant, B. A. Leland, D. R. Henry and J. G. Nourse, J. Chem. Inf. Comput. Sci., 2002, 42, 1273–1280 CrossRef CAS PubMed.
- G. E. Hinton and S. Roweis, Adv. Neural Inf. Process Syst., 2002, 15, 857–864 Search PubMed.
- L. Van der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra06022a |
| ‡ Sheng Li and Xinqiao Wang contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |