Open Access Article
Open Access ArticleUltrasensitive detection of antimicrobial resistance genes using hybridization chain reaction employing carbon dots†
Harshil Thakkara,
Sonal Thakore *a,
Manoj Baghelb,
Sanjay Kosarab,
Heli Upadhyayab,
Hemanta Chalanb and
Devarshi Gajjar*b
*a,
Manoj Baghelb,
Sanjay Kosarab,
Heli Upadhyayab,
Hemanta Chalanb and
Devarshi Gajjar*b
aDepartment of Chemistry, Faculty of Science, The Maharaja Sayajirao University of Baroda, 390002, India. E-mail: chemistry2797@yahoo.com; sonal.thakore-chem@msubaroda.ac.in
bDepartment of Microbiology and Biotechnology Centre, Faculty of Science, The Maharaja Sayajirao University of Baroda, 390002, India. E-mail: devarshi.gajjar-microbio@msubaroda.ac.in
First published on 10th December 2024
Abstract
One of the top 10 global concerns include AntiMicrobial Resistance (AMR), which warrants the need to develop materials and methods for detection of AMR genes. Here, we propose a proof-of-concept approach for selective and ultrasensitive detection of AMR gene employing fluorescent carbon dots. Waste pistachio shell derived green emissive carbon dots (PCDs) with a high quantum yield of 24 were prepared via hydrothermal carbonization process and characterised using microscopic and spectroscopic techniques. The fluorescence-based Hybridization Chain Reaction (HCR) mediated sensing studies demonstrated the ability of the PCD sensor to detect AMR gene, compared to random and single mismatch DNA with a limit of detection of 16.17 pM. This strategy of waste valorization to design fluorescent probe offer excellent cost-effective and sustainable alternative for ultra-trace level detection of DNA.
Introduction
Antimicrobial resistance (AMR) is a phenomenon when bacteria, viruses, fungi and parasites no longer respond to antimicrobial medicines. The natural process of AMR, is a phenomenon to which the public health crisis has increased due to inappropriate use of antibiotics than the phenomenon itself. Antibiotic resistance genes (ARGs) are categorized under global priority list in 2017 by the World Health Organization (WHO), significantly accelerating in the past decade in all environments, including natural, engineered, and clinical habitats. Among these ARGs, carbapenem resistance of Gram-negative bacteria is of major concern.1,2 The resistance to carbapenem by Klebsiella pneumoniae can be attributed to occurrence of isolates with lactamase blaKPC, metallolactamase blaNDM, and oxacillinase blaOXA-48, with blaOXA-48 and blaNDM1/5 being the most common ones.3Researchers have recently found that nucleic acids hold great potential for applications in medicine. These possess significant properties like programmable base-pairing, well-determined structure, high stability and flexibility. DNA is known to be a self-assembling building block and a flexible entity to construct dynamic and automatic nanoscale switchable devices due to metastable configurations in a dynamic environment. These superior properties and functions demonstrate the importance of nucleic acid tests (NATs) based on the principles of RNA transcription and intracellular DNA replication.4,5 Toehold-Mediated Strand Displacement (TMSD) is one of the NATs, where the metastable secondary structures of nucleic acid species is determined by an isothermal polymerization in which one strand displaces a second in contact with a third which is complementary to both leading to replication. Hybridization chain reaction (HCR) is an enzyme-free simple and efficient isothermal nucleic acid amplification technique, with exciting features and bright prospects in biosensing and thus has been intensively investigated in the past decade.6,7 With advantages like great assembly kinetics, facile operation, and an enzyme-free and isothermal reaction, it is much suitable for the analysis of temperature-sensitive targets like proteins, exosomes, nucleic acids and cells.8,9 The reaction process is simplified with mild conditions, thus good practical use is verified. In the classic mechanism of the HCR, the DNA target (T-DNA) will trigger the cooperative cross-opening of two DNA hairpin probes leading to amplification. Due to the absence of requirements of an enzyme, isothermal amplification, excellent sensitivity, and adaptable structures, it has been coupled with multiple detection strategies. These include colorimetric,10,11 fluorimetric,12 chemiluminescence-based, and electrochemical13 signal readout methods to detect analytes sensitively. It therefore provides a significant advantage in the development of potential molecular instruments and methods for biosensing, bioimaging, and biomedicine as a substitute for conventional nucleic acid amplification techniques, such as polymerase chain reaction (PCR),14 loop-mediated isothermal amplification (LAMP),15 helicase dependent amplification (HDA),16 strand displacement amplification (SDA),17 and rolling circle amplification (RCA).18
Several materials and methods have been developed so far to detect DNA using HCR. The basic principle behind the optical detection approach so far is based on the interaction between fluorophore and quencher, where the fluorophore tagged hairpin probes (HP) interact with the quencher in absence of T-DNA. Presence of T-DNA detaches the probes, leading to fluorescence recurrence. There have been several reports including Au nanoparticles,11 graphene oxide quantum dots,12 MoS2 sheets,19 pyrene-labelled hairpins20 and 2D-TPA-COF nanosheets21 for HCR mediated DNA detection. Most of these involve materials that require tedious, cost intensive and time-consuming synthesis. These materials require fluorophore tagged HPs which are quite expensive22 and some of them have potential cytotoxic effects. Thus, there is a need for inexpensive and sensitive materials and methods for DNA detection which can be translated for practical applications.
Carbon based quantum dots (CQDs) are a new class of fluorescent nanomaterials that have gained attention as sensors due to their remarkable properties like low cost, facile synthesis, high water solubility, excellent photostability, large surface area, surface functionalities, and biocompatibility.23 Further, CQDs demonstrate selective sensing ability towards metal ions and various molecules,24 and hence are recognized for their applications in optoelectronics and bioimaging.25 These remarkable photophysical properties, coupled with their applications make them effective alternatives to organic dyes, semiconductor quantum dots, and polymer dots.26 CQDs are synthesized by chemical or physical methods, using synthetic and biogenic precursors. Abundant bioactive molecules are present in biowastes generated worldwide. Owing to this, biogenic synthesis offers an excellent approach of synergistically converting the waste into functional materials for selective and ultra-sensitive optical detection. Pistachio is a commercially available product belonging to the cashew family. The hard outer shell of pistachio contains cellulose, hemicellulose and lignin. These carbon-rich hard shells are slow-decomposing, and can take several years to fully degrade in a compost pile. We propose rational valorization of these carbon rich pistachio shells to develop fluorescent CDs which could be useful for rapid, selective and ultra-sensitive detection of anti-microbial resistance genes. To the best of our knowledge, this is the first report based on HCR mediated detection of DNA using fluorescent carbon dots.
Results and discussion
One pot synthesis of pistachio shell based carbon dots (PCDs)
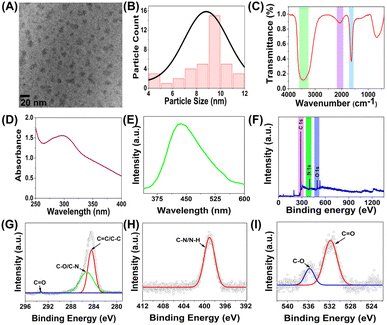
Considering the abundant lignocellulosic waste generated from nuts, we propose an environmentally benign hydrothermal treatment method to convert waste pistachio shells into fluorescent carbon dots (PCDs) as represented in Fig. 1. The synthesis was done at optimized conditions of 150 °C for 6 hours, as represented in Fig. S4.† The active molecules, especially lignocellulosic material present in the shell undergo dehydration, hydrolysis, hydrothermal oxidation and polycondensation followed by nuclear burst and carbonization to form PCDs.27 The formation mechanism of PCDs from the lignin present in pistachio shell is represented in figure. As per the literature reports Nitrogen dopants are known to enhance the PL emission24 and also provide partial surface positive charge, similar to that observed on histone proteins, facilitating interaction with negatively charged DNA.28 Considering these, PCDs were doped with Nitrogen using ethylene diamine.Characterization
Morphology and size of the PCDs was determined using HRTEM images. Fig. 2A reveal spherical PCDs with average particle diameter of 9.4 nm as observed in Fig. 2B. The microstructure and the functional group analysis were done using FTIR, shown in Fig. 2C. The FTIR spectra demonstrated a broad peak of carboxyl O–H and N–H stretching at 3433 cm−1. A peak at 1643 cm−1 for carbonyl stretching was observed. These results confirm the presence of –COOH, –OH and –NH2 functionalities on the surface of PCDs. Another peak was observed at 2076 cm−1 due C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching, formed during the dehydration process during PCD formation. These functional group attributes the PCDs with the exceptional water dispersibility and stability in aqueous medium. The size distribution of PCDs, obtained via DLS method (Fig. S5(A)†), correlates with the HRTEM images. UV-Vis (Fig. 2D) and PL (Fig. 2E) spectroscopic studies revealed absorbance and emission at 300 nm and 420 nm respectively. The synthesized PCDs showed a QY of 24, which is much higher than that of the previously synthesized biogenic CDs. The absorbance at 300 nm corresponds to n–π* transitions in the fluorophores present in PCDs. XPS spectra was obtained to analyse the surface stated of PCDs. Fig. 2F represents the survey spectra, which depict peaks at 284.4 eV, 398.7 eV and 531.8 eV for C1s, N1s and O1s respectively on the surface of PCDs. Deconvoluted spectra of C1s (Fig. 2G) exhibit peaks at 284.3 eV, 285.3 eV and 293 eV corresponding to C–C/C
C stretching, formed during the dehydration process during PCD formation. These functional group attributes the PCDs with the exceptional water dispersibility and stability in aqueous medium. The size distribution of PCDs, obtained via DLS method (Fig. S5(A)†), correlates with the HRTEM images. UV-Vis (Fig. 2D) and PL (Fig. 2E) spectroscopic studies revealed absorbance and emission at 300 nm and 420 nm respectively. The synthesized PCDs showed a QY of 24, which is much higher than that of the previously synthesized biogenic CDs. The absorbance at 300 nm corresponds to n–π* transitions in the fluorophores present in PCDs. XPS spectra was obtained to analyse the surface stated of PCDs. Fig. 2F represents the survey spectra, which depict peaks at 284.4 eV, 398.7 eV and 531.8 eV for C1s, N1s and O1s respectively on the surface of PCDs. Deconvoluted spectra of C1s (Fig. 2G) exhibit peaks at 284.3 eV, 285.3 eV and 293 eV corresponding to C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–H, C–N/C–O and C
C/C–H, C–N/C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O surface states.23 N1s spectra (Fig. 2H) demonstrated peak at 399.1 eV representing 1°/2° amino nitrogen (C–N/N–H).29 The peaks at 531.3 eV and 535.7 eV in the O1s spectra can be attributed to O–H/C–O and C
O surface states.23 N1s spectra (Fig. 2H) demonstrated peak at 399.1 eV representing 1°/2° amino nitrogen (C–N/N–H).29 The peaks at 531.3 eV and 535.7 eV in the O1s spectra can be attributed to O–H/C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O–C
O/O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O states23 (Fig. 2I).
O states23 (Fig. 2I).
Sensing studies
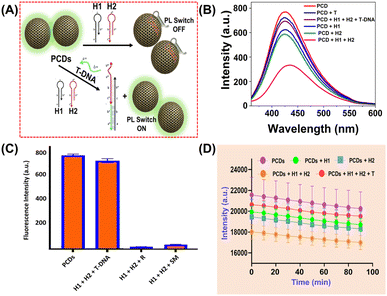
ssDNA is known to interact with surface functional groups on the surface of CQDs via supramolecular and electrostatic interactions, eventually causing PL quenching of CQDs (ref). This principle inspired us to develop a proof-of-concept optical method for specific AMR DNA detection. The method does not employ cost-intensive fluorophore tagged HPs22 due to the inherent PL emission of PCDs. Interaction of single stranded toe-hold regions of HPs with carbon dots via aforementioned interactions in absence of T-DNA caused PL quenching. But in the presence of specific T-DNA, the HPs detached from the PCD surface due to their enhanced thermodynamic affinity with the target, eventually leading to PL recurrence. Till date, no such report on DNA detection using carbon dots via HCR method has been reported. The sensing studies involve four steps: (A) optimization of PL quenching using HPs of different toe-hold length, (B) validation of the proposed detection method using optimized HPs, (C) extrapolation of the proof-of-concept detection approach for AMR NDM gene detection and (D) to determine the sensitivity of sensor towards varying NDM gene concentrations. (A) Optimization of HPs. The binding of the oligos H1 and H2 on the PCD surface were compared via PL quenching experiments with 3 different sets of HPs with different toe-hold length (first set of 15 bp and 10 bp, second set with 10 bp each and third set of 6 bp each). The sequences of the oligos and their report is represented in Tables S1 and S2† respectively. The studies as depicted in Fig. S8,† demonstrated significant PL quenching with HPs with 10 bp toe-hold length compared to other two sets. According to Ang et al. and He et al., the toehold region greater than 12 nucleotide bases leads to secondary structure formation,30,31 hence demonstrating lower binding affinities with PCDs. On the contrary, HPs with 6 bp toe-hold region could not lead to sufficient supramolecular and electrostatic interaction with PCDs, hence demonstrating minimal PL quenching. Considering these studies, AMR NDM gene with 10 bp toe-hold length was designed and used for further studies. (B) Validation of proof-of-concept optical detection. The PL quenching and recurrence experiments of PCDs were performed using optimized HP sequences. The studies, represented in Fig. S9† depicted that the PCDs interacted selectively with H1 and H2, compared to T-DNA leading to enhanced PL quenching and decrease in PL intensity. This can be attributed to the longer length of the T-DNA which leads to formation of secondary structures and size compatibility between the PCDs and target. Further, studies performed by injection of T-DNA int the solution containing PCDs, H1 and H2 demonstrated amplification and PL recurrence, which is primarily due to greater thermodynamic affinity of the HPs to T-DNA compared to PCDs. (C) Detection of AMR NDM gene. Increasing concern of AMR genes and significant results from the validation experiments inspired us to extrapolate the studies for detection of NDM gene as per mechanism shown in Fig. 3A. Qualitative studies were done in two parts to determine the selectivity of PCDs. The first set of studies were performed to establish selective binding of HPs compared to target NDM (NDM_T) gene as demonstrated in Fig. 3B. Results were observed similar to that of (B) viz., NDM_H1 and NDM_H2 depicting significant PL quenching compared to T-DNA. Real time monitoring studies were performed to determine the optimum interaction time to observe PL quenching and recurrence. Fig. 3D, showed similar sequential PL switch OFF and ON results. Real time studies validated rapid and consistent interactions as per the proof-of-concept optical detection. The second set of studies were performed to confirm selective PL recurrence of PCDs in presence of target compared to single mismatch (NDM_SM) and random (NDM_R) as depicted in Fig. 3C. The studies revealed selective fluorescence switch ON, specifically with T-DNA compared to NDM_R and NDM_SM. This is primarily due to selective HCR amplification in presence of T-DNA.21 (D) Sensitivity studies. To evaluate the sensitivity of PCD sensors for specific NDM gene detection, PL quenching studies were performed with varying concentration of NDM_T in the range of 0–1 μM. The studies revealed increase in PL recurrence upon increasing T-DNA concentration as observed in Fig. 4A. The Stern–Volmer graph of (F0/F − 1) versus the DNA concentration in 0–1 μM range depicted good linearity with R2 value of 0.998 (Fig. 4B). The results showed LOD of 16.17 pM which is much lower than other reports as summarized in Table S2.†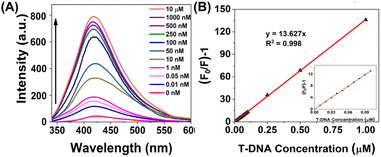 | ||
| Fig. 4 (A) Graph showing PL recurrence with different T-DNA concentrations and (B) Stern–Volmer plot showing PL switch ON. | ||
Conclusions and future perspectives
To summarise, we report the synthesis and characterization of pistachio shell derived carbon dots via low temperature hydrothermal carbonization process. As a proof-of-concept application, the PCDs were used as a novel optical sensing platform for DNA detection with excellent selectivity and high sensitivity. Thorough studies were performed to establish the concept, which was further extrapolated for picomolar level detection of globally concerned anti-microbial resistance genes. Through this, we envisage a strategy to valorise waste to design and synthesize cost-effective and non-toxic carbon-based quantum dots with inherent fluorescence for rapid ultra-sensitive detection of DNA using economical non-fluorophore tagged HCR probes. The work opens up a new promising strategy for clinical and diagnostic applications.Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to DST-SERB (SPG/2021/0003149) and GSBTM (Grant Number: GSBTM/JD(R&D)/616/21-22/1236) for financial assistance. The authors acknowledge MNIT Jaipur for XPS Analysis and ICON Labs, Mumbai for HRTEM.References
- Z. Zhang, Q. Zhang, T. Wang, N. Xu, T. Lu, W. Hong, J. Penuelas, M. Gillings, M. Wang, W. Gao and H. Qian, Nat. Commun., 2022, 13, 1553 CrossRef CAS
.
- C. Biondo, Pathogens, 2023, 12, 0–1 Search PubMed
.
- H. Zou, B. Berglund, S. Wang, Z. Zhou, C. Gu, L. Zhao, C. Meng and X. Li, Environ. Pollut., 2022, 306, 1–9 CrossRef
.
- S. F. Khan, P. Rathod, V. K. Gupta, P. B. Khedekar and R. V. Chikhale, Anal. Chem., 2024, 96, 8124–8146 CrossRef CAS
.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed
.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC
.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS
.
- Y. Jiang, X. Chen, N. Feng and P. Miao, Anal. Chem., 2022, 94, 14755–14760 CrossRef CAS
.
- M. Y. Wang, W. J. Jing, L. J. Wang, L. P. Jia, R. N. Ma, W. Zhang, L. Shang, X. J. Li, Q. W. Xue and H. S. Wang, Biosens. Bioelectron., 2023, 226, 115116 CrossRef CAS
.
- C. Zhang, J. Lou, W. Tu, J. Bao and Z. Dai, Analyst, 2015, 140, 506–511 RSC
.
- D. Yuan, X. Fang, Y. Liu, J. Kong and Q. Chen, Analyst, 2019, 144, 3886–3891 RSC
.
- L. Chen, L. Song, Y. Zhang, P. Wang, Z. Xiao, Y. Guo and F. Cao, ACS Appl. Mater. Interfaces, 2016, 8, 11255–11261 CrossRef CAS
.
- H. Li, S. Li, H. Li, X. Li, M. Zhu and F. Xia, Anal. Chem., 2021, 93, 8354–8361 CrossRef
.
- E. F. Remmers, M. J. Ombrello and R. M. Siegel, Principles and Techniques in Molecular Biology, Elsevier Ltd, 6th edn, 2015, vol. 1–2 Search PubMed
.
- F. Azevedo-Nogueira, P. Martins-Lopes and S. Gomes, Crop Prot., 2020, 132, 105106 CrossRef CAS
.
- M. Gavrilov, J. Y. C. Yang, R. S. Zou, W. Ma, C. Y. Lee, S. Mohapatra, J. Kang, T. W. Liao, S. Myong and T. Ha, Nat. Commun., 2022, 13, 1–14 Search PubMed
.
- M. Mohammadniaei, M. Zhang, J. Ashley, U. B. Christensen, L. J. Friis-Hansen, R. Gregersen, J. G. Lisby, T. L. Benfield, F. E. Nielsen, J. Henning Rasmussen, E. B. Pedersen, A. C. R. Olinger, L. T. Kolding, M. Naseri, T. Zheng, W. Wang, J. Gorodkin and Y. Sun, Nat. Commun., 2021, 12, 1–12 CrossRef
.
- M. G. Mohsen and E. T. Kool, Acc. Chem. Res., 2016, 49, 2540–2550 CrossRef CAS
.
- J. Huang, L. Ye, X. Gao, H. Li, J. Xu and Z. Li, J. Mater. Chem. B, 2015, 3, 2395–2401 RSC
.
- Z. He, J. Wu, B. Qiao, H. Pei, Q. Xia, Q. Wu and H. Ju, ACS Appl. Bio Mater., 2020, 3, 5342–5349 CrossRef CAS
.
- Y. Peng, Y. Huang, Y. Zhu, B. Chen, L. Wang, Z. Lai, Z. Zhang, M. Zhao, C. Tan, N. Yang, F. Shao, Y. Han and H. Zhang, J. Am. Chem. Soc., 2017, 139, 8698–8704 CrossRef CAS PubMed
.
- L. Song, Y. Zhang, J. Li, Q. Gao, H. Qi and C. Zhang, Appl. Spectrosc., 2016, 70, 688–694 CrossRef CAS
.
- H. Thakkar, P. Bhandary and S. Thakore, ACS Appl. Nano Mater., 2023, 6, 16253–16266 CrossRef CAS
.
- M. Das, H. Thakkar, D. Patel and S. Thakore, J. Environ. Chem. Eng., 2021, 9, 106312 CrossRef CAS
.
- H. Li, Z. Kang, Y. Liu and S. T. Lee, J. Mater. Chem., 2012, 22, 24230–24253 RSC
.
- M. L. Liu, B. Chen and M. Li, Green Chem., 2019, 449–471 RSC
.
- X. Wang, Y. Feng, P. Dong and J. Huang, Sustainable Mater. Technol., 2019, 7, 1–9 Search PubMed
.
- F. Li, Q. Cai, X. Hao, C. Zhao, Z. Huang, Y. Zheng, X. Lin and S. Weng, RSC Adv., 2019, 9, 12462–12469 RSC
.
- M. Liu, Y. Xu, F. Niu, J. J. Gooding and J. Liu, Analyst, 2016, 141, 2657–2664 RSC
.
- Y. S. Ang and L. Y. L. Yung, Chem. Commun., 2016, 52, 4219–4222 RSC
.
- H. He, J. Dai, Z. Duan, B. Zheng, Y. Meng, Y. Guo and D. Xiao, Sci. Rep., 2016, 6, 1–7 CrossRef
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, Fig. S1–S9, and Tables S1–S3 are available for this paper. See DOI: https://doi.org/10.1039/d4ra07517j |
| This journal is © The Royal Society of Chemistry 2024 |